
Are you running an RNAi experiment?
If so, don't forget about using controls! The last thing you want is a result attributed to an off-target gene knockdown, or an effect due to experimental manipulation unrelated to RNAi. Critical to any gene silencing or microRNA modulation experiment, RNAi controls are essential for accurate interpretation of results. A carefully designed RNAi experiment should minimally include untreated cells, mock transfected cells, a negative and a positive control. Let's take a closer look. Always include the following controls to cover all your bases:
Untreated Cells
Untreated cells are the normal population against which all other samples may be compared. Untreated cells determine the baseline level of cell viability, phenotype, and target mRNA gene expression levels.
Mock Transfection
Treatment of cells with only the delivery agent (e.g. transfection reagent alone or empty vector) to control for cellular effects due to transfection reagent exposure or transduction. This control is important because transfection reagents or transduction can easily alter gene expression.
Non-targeting Negative Control
A negative control should be an RNAi reagent with a non-targeting sequence to provide a baseline reference for target-specific knockdown and distinguish sequence-specific silencing from non-specific effects. Ideally this control should be used under the same conditions (concentration, delivery method, duration of knockdown, and detection methods) as the targeted RNAi and have no effect on cell viability, phenotype or target mRNA and protein levels. Non-targeting controls should be validated, randomly scrambled controls perform very poorly.
Positive Control
This is a validated control to monitor efficiency of delivery across experimental replicates, conditions, or cell types. The best positive control target is an endogenous housekeeping gene, such as Cyclophilin B, that is under normal transcriptional regulation, whose expression does not fluctuate with the cell cycle in the cell line being tested, and whose knockdown does not affect the cell phenotype or viability.
Fluorescent Control
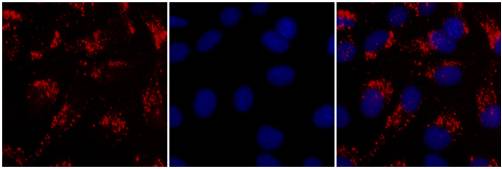
This is an optional, but helpful visual control that evaluates transfection efficiency. The fluorescence may be detected by either fluorescence microscopy or flow cytometry. Don't forget that using conventional fluorescent labeled siRNA or shRNA does not correlate quantitatively with gene knockdown, only with delivery efficiency.
Once the appropriate controls have been identified for the experimental system being used, you can move on to optimizing your experimental conditions and have confidence in the interpretation of your RNAi results!
Browse Horizon's siRNA controls
To learn more read our informative application note
Questions or comments?
