Prime editing is a CRISPR-based method for introducing small genetic changes without making a double-stranded break (DSB). By substantially modifying the architecture of the guide RNA, along with fusing a reverse transcriptase to a Cas9 nickase, Anzalone, Liu and colleagues have generated an editing system that can introduce single base-pair swaps, substitute multiple different bases within a small target window, or generate small insertions and deletions, with precision that could outweigh standard CRISPR-Cas gene editing.
Standard CRISPR-Cas9 techniques induce gene editing by using a guide RNA to traffic the Cas9 nuclease to a specific region of the genome where it creates a DSB, which is repaired in a mammalian cell by one of two (or more) distinct mechanisms: ‘error-prone’ nonhomologous end-joining (NHEJ) and ‘error-free’ homology-directed repair (HDR). Most of the time, NHEJ and other imperfect ligation outcomes result in insertions and deletions (indels), which can cause gene disruption and knockout. A specific edit can be made with HDR by the inclusion of a DNA template, but this outcome is often inefficient and can be difficult to find in a mixed population of cells, where NHEJ repaired cells and remaining wild-type cells will form the majority.
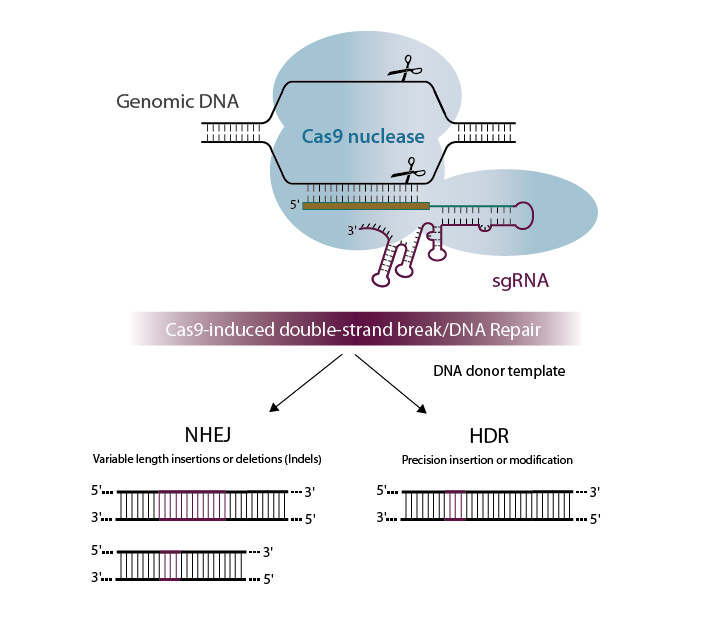
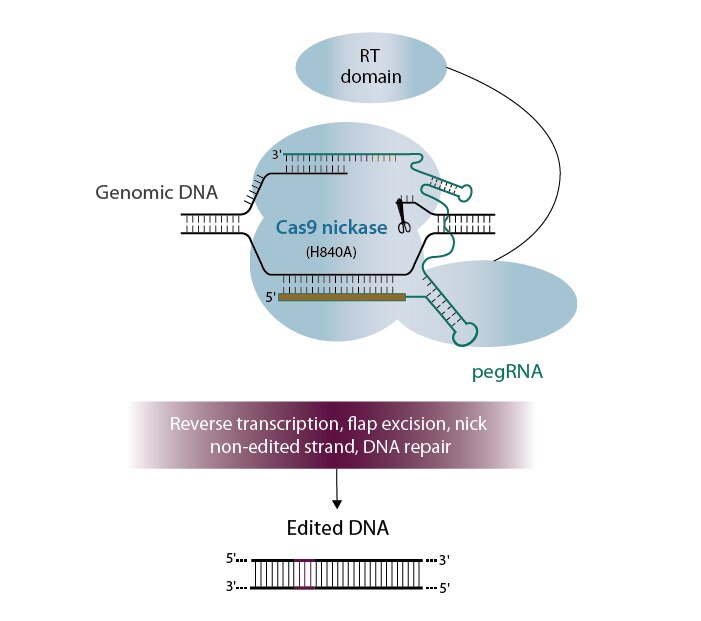
Prime editing (Anzalone, 2019) uses a Cas9 nickase fused to an engineered reverse transcriptase (RT). This fusion is used with a modified guide RNA that is lengthened to contain both a RT primer binding site and RNA template containing the desired edit; this entire guide complex is known as a PRIME editing guide RNA or pegRNA. The combination of the pegRNA and the nickase Cas9-RT fusion influences the DNA repair mechanisms in a cell to incorporate the edit into the desired location by copying the RNA template and substituting this into the target site. The target DNA is only nicked and is not cut on both strands at the same location, thereby limiting indels to only a small fraction of the edits, versus a vast majority of the outcomes when a DSB is made.
Currently, Prime editing does appear to be a sharper knife in the toolbox and is effective in the context of a dual plasmid system containing the nickase Cas9-RT fusion and the pegRNA plasmid. However, synthetic pegRNAs are on the Horizon (pun intended) and could allow for more efficient and cleaner editing. The use of synthetic guides enables easier delivery and transient expression of the guide RNA. Our custom synthesis capabilities allow synthesis of long RNA for any application with multiple modifications and processing options available. Prime editing is the latest application to take advantage of our long RNA capabilities.
Request a quote for your long RNA or contact us and we will help get your project started!
