A lentiviral vector permitting temporal control of Cas9 expression helps reveal the function of essential genes, while avoiding constitutive expression of Cas9.
- Temporal control of Cas9 expression and gene editing
- Facilitates enrichment of edited cells
- High precision and improved experimental flexibility

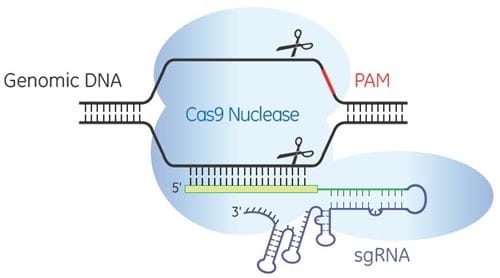
The Edit-R™ Inducible Lentiviral Cas9 Expression Vector is based on one of two critical components of the CRISPR-Cas9 system required for gene editing in mammalian cells – the Cas9 nuclease enzyme. Used in conjunction with an Edit-R Lentiviral sgRNA designed to target the gene of interest, the Edit-R Inducible Lentiviral Cas9 Expression Vector provides researchers with temporal control of gene knockout using an optimized doxycycline inducible system.
Temporal control of Cas9 expression and gene editing
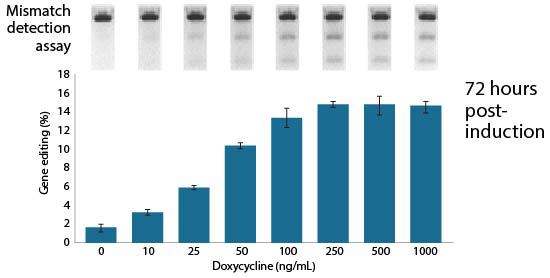
The Edit-R Inducible Lentiviral Cas9 Expression Vector contains a human codon-optimized version of the Cas9 gene under the control of the highly responsive Tet-On® 3G doxycycline inducible promoter. This allows gene knockouts to be obtained by treatment with an optimized concentration of doxycycline at a precisely defined time. As well as permitting tight regulation of Cas9 expression with minimal basal expression, the Edit-R Inducible Lentiviral Cas9 Expression Vector provides potent activation upon induction, even at very low doses of doxycycline.
Facilitates enrichment of edited cells
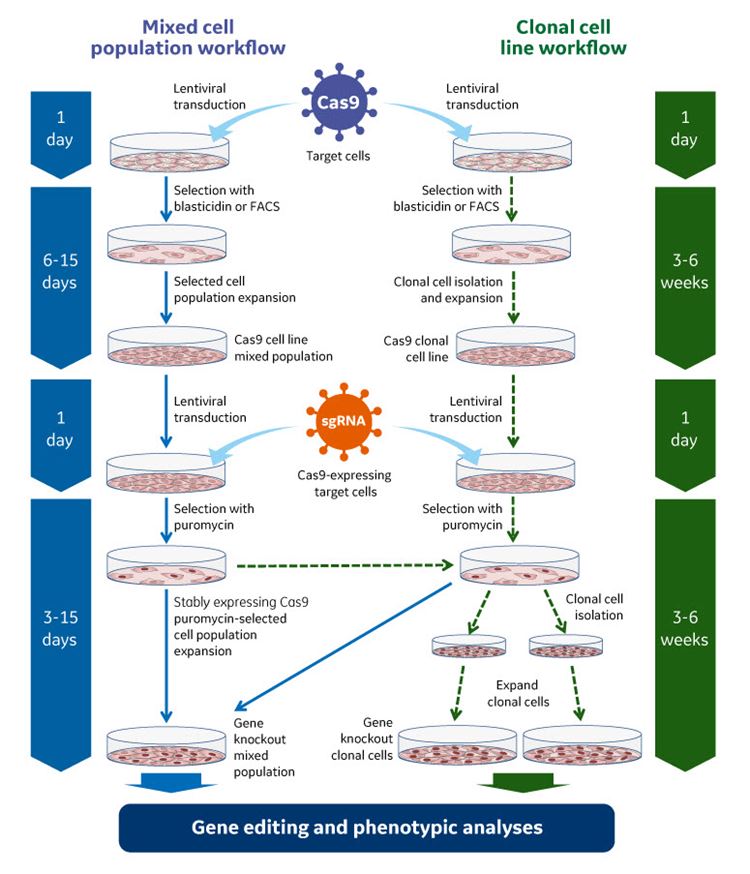
By constitutively expressing the blasticidin resistance gene (BlastR), the Edit-R Inducible Lentiviral Cas9 Expression Vector enables selection of Cas9-expressing cells prior to sgRNA transduction. In turn, this increases the percentage of gene edited cells available for downstream studies for identifying functional knockouts.
High precision and improved experimental flexibility
The Edit-R Inducible Lentiviral Cas9 Expression Vector provides robust expression of the Cas9 nuclease when induced, and minimal promoter transcription in the off state for high precision in gene editing. It can be used in conjunction with an Edit-R Lentiviral sgRNA targeting any gene of interest, affording maximum flexibility.

Written by Ryan Donnelly
He holds a Professional Science Masters in Molecular Biotechnology from George Washington University and Bachelors in Chemistry from the University of Florida.
Ready to get started with inducible lentiviral Cas9 expression? See our product page.
To learn more about the specifics of the product. Download the technical manual.
Do you have further questions or need some help troubleshooting your experiment, contact our Scientific Support team anytime.
