The prospect of gene editing in human primary cells has enabled the study of gene function in a context beyond immortalized cell lines and has driven the discovery of immune-based therapeutic applications. However, CRISPR-Cas9 gene editing is notoriously difficult in many primary human cell types. When applied to human myeloid cells, which play an important role in pathogen response, autoinflammatory disease, and cancer malignancy, gene editing has been particularly inefficient, until now.
The labs of Alex Marson, MD, PhD, and Nevan Krogan, PhD, which reside inside the Gladstone Institute of UCSF, developed a protocol using Horizon Discovery’s Edit-R™ synthetic guides to successfully edit genes inside monocytes, all the while preserving their functionality and differentiation capabilities. Using Cas9 ribonucleoprotein (RNP) complexes, the labs set out to optimize nucleofection conditions that balanced editing efficiency, cell morphology, and cell survival. Accomplishing this required generation of novel cell differentiation and culturing methods, along with fine-tuning instrumentation parameters.
The authors showcase that Horizon’s SAMHD1 synthetic guide RNA delivered as a Cas9-RNP complex in primary human CD14+ monocytes results in robust SAMHD1 gene knockout that persists in subsequently differentiated monocyte-derived macrophages (MDMs) or monocyte-derived dendritic cells (MDDCs). In addition, a dramatic ablation of SAMHD1-associated viral restriction factor function clearly demonstrates that this platform is suitable for functional and phenotypic assessment such as phagocytic ability and cell-type specific marker gene expression.
See a snapshot of the figures from the Cell Reports publication here:
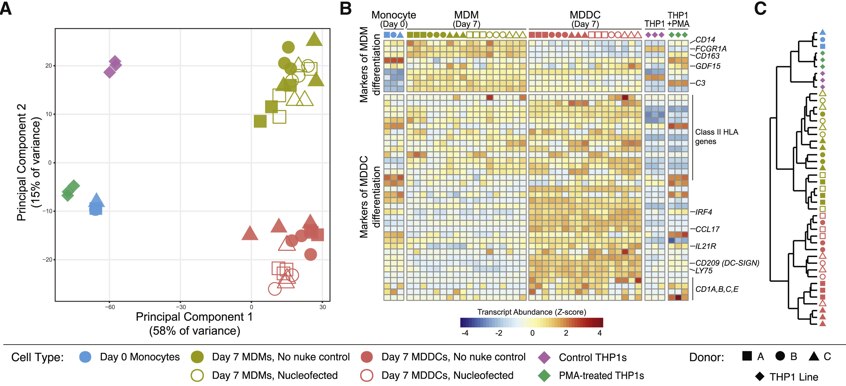
Figure 2. CRISPR-Cas9-mediated gene knockout preserves key aspects of differentiation and function in targeted myeloid cells.(A) Principal-component analysis of RNA sequencing (RNA-seq) from the indicated cell types. (B) Normalized transcript abundance (Z score) for selected markers of MDM or MDDC differentiation (Lehtonen et al., 2007). (C) Dendrogram of hierarchical clustering of the data in (B) by Euclidean distance.
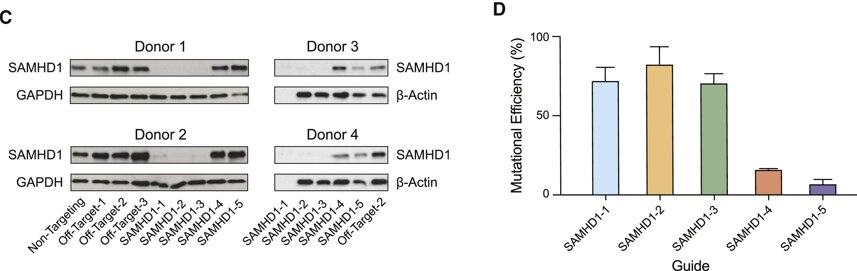
Figure 3. Generation of isogenic monocyte-derived macrophages for functional evaluation of an HIV-1 host restriction factor. (Parts C and D) Quantification of SAMHD1 knockout by immunoblot (C) and sequencing (D). No protein sample was available for guide SAMHD1-1 in donors 3 and 4; Sanger sequencing was analyzed for mutational efficiency by TIDE, bars represent means ± SDs for at least 3 biological replicates.
This novel methodology is an important step enabling research in primary human monocytes, allowing for robust knockout and functional and phenotypic investigation. The ability to edit genes in these cells will open the door for new immunological and auto-immunological response treatments, and potentially accelerate the development of myeloid cell-based immune-oncology therapies.
 Written by Ashleigh Keller
Written by Ashleigh Keller
Ashleigh joined the R&D team as a Scientist I at Horizon Discovery in January 2020. Her work has contributed to the development and application of CRISPR gene editing tools, most notably the CRISPRmod portfolio of reagents that promote (CRISPRa) or repress (CRISPRi) target gene transcription. Ashleigh obtained her Bachelor of Science from Colorado State University and is currently working towards a Master’s degree in Microbiology and Biochemistry from the University of Florida.
Read the full publication in Cell Reports
Efficient generation of isogenic primary human myeloid cells using CRISPR-Cas9 ribonucleoproteins Hiatt, J., et al. Cell Reports. 35:6 109105. https://doi.org/10.1016/j.celrep.2021.109105
