Introducing the new ready-to-use CRISPRmod CRISPRi All-in-one Lentiviral sgRNA Whole Genome Pooled Library! The library uses the dCas9-SALL1-SDS3 repressor developed in house1, which has been shown to give greater and longer lasting repression compared to the standard dCas9-KRAB repressor when used with synthetic sgRNAs. In this blog post, we compare this library to a similar library using dCas9-KRAB.
An all-in-one CRISPRi construct for faster results
The CRISPRi All-in-one Lentiviral Pooled Library uses a single lentiviral vector for expression of the CRISPRi repressor (dCas9-SALL1-SDS3) and the gene-specific sgRNA. This means that only one lentiviral transduction is necessary, so that you can achieve faster results and streamline your workflow with a single round of antibiotic selection (Figure 1).

Figure 1: Streamlined workflow with the All-in-one CRISPRi library
Standard screening workflow using the CRISPRmod CRISPRi All-in-one Lentiviral Pooled Library platform, here performed in A375 melanoma cells treated with vemurafenib (PLX-4032) for 14 days following transduction and puromycin selection. Genomic DNA was then isolated from the reference and experimental populations of transduced cells and sgRNA constructs within the isolated gDNA were amplified and prepared for high-throughput sequencing on an Illumina platform. The integrated sgRNA sequences in both reference and experimental samples were identified and relative abundance compared.
Increased essential gene dropout with the CRISPRi All-in-one library
To compare our All-in-one Lentiviral Pooled Library to a similar library using the dCas9-KRAB repressor1, A375 cells were transduced with one of the two libraries, then vemurafenib (a drug which causes programmed cell death in melanoma cell lines) was applied as a selective pressure (Figure 1).
We then analyzed 684 core essential genes in each screen. More of the sgRNAs targeting essential genes (purple in Figure 2) dropped out when using the dCas9-SDS3-SALL1 repressor, indicating greater repression with dCas9-SDS3-SALL1 than with dCas9-KRAB. Analysis of non-essential genes (green in Figure 2) showed neutral behavior in both screens, since knockdown of these genes did not affect cell survival.

Figure 2. Enhanced essential gene dropout with the CRISPRmod CRISPRi All-in-one library
Guide level analysis of a published set of 684 core essential genes (CEG, purple) and non-essential genes (NEG, green) at the end of the screen as compared the start2 shows enhanced essential gene dropout with the CRISPRmod CRISPRi All-in-one lentiviral sgRNA Whole Genome Pooled Library (dark purple/green) compared to a design-matched dCas9-KRAB whole genome pooled library (light purple/green).
Highly concordant hits between the dCas9-SALL1-SDS3 and dCas9-KRAB libraries
The screens using the dCas9-SALL1-SDS3 and dCas9-KRAB libraries both independently identified many of the same hits (shown in orange in Figure 3), validating the effectiveness of our dCas9-SALL1-SDS3 All-in-one system for CRISPRi screens. Out of the top 20 hits from each screen, 11 of them were the same (Figure 4).
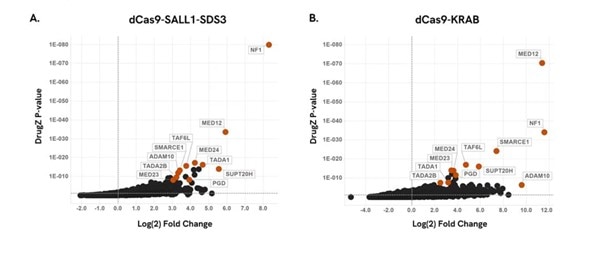
Figure 3. Highly concordant hits between the dCas9-SALL1-SDS3 and dCas9-KRAB systems
CRISPRmod CRISPRi Human All-in-one lentiviral sgRNA Whole Genome Pooled Library identifies highly concordant resistant hits in A375 melanoma cells treated with vemurafenib (PLX-4032). DrugZ screen analysis showing log2 fold enrichment of each gene and associated p-values; significantly enriched genes (positive Log2 Fold Change) drive resistance to the drug treatment indicating these genes potentially act as drug antagonists. Highlighted genes were also identified as top resistance hits in a screen performed with a single vector dCas9-KRAB whole genome library.
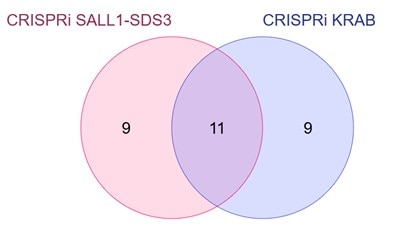
Figure 4: Concordance of the top 20 hits between whole-genome CRISPRi screens performed using the CRISPRmod All-in-one dCas9-SALL1-SDS3 and dCas9-KRAB systems
Ready to take your research to the next level?
Get started with the CRISPRmod CRISPRi All-in-one Lentiviral Whole Genome Library for more efficient CRISPRi screens. Explore the details including protocols and workflows.
References
- le Sage C, Lawo S, Panicker P, Scales TME, Rahman SA, Little AS, McCarthy NJ, Moore JD, Cross BCS. Dual direction CRISPR transcriptional regulation screening uncovers gene networks driving drug resistance. Sci Rep. 2017 Dec 18;7(1):17693. doi: 10.1038/s41598-017-18172-6. PMID: 29255251; PMCID: PMC5735189.
- Hart T, Tong AHY, Chan K, Van Leeuwen J, Seetharaman A, Aregger M, Chandrashekhar M, Hustedt N, Seth S, Noonan A, Habsid A, Sizova O, Nedyalkova L, Climie R, Tworzyanski L, Lawson K, Sartori MA, Alibeh S, Tieu D, Masud S, Mero P, Weiss A, Brown KR, Usaj M, Billmann M, Rahman M, Constanzo M, Myers CL, Andrews BJ, Boone C, Durocher D, Moffat J. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3 (Bethesda). 2017 Aug 7;7(8):2719-2727. doi: 10.1534/g3.117.041277. PMID: 28655737; PMCID: PMC5555476.
