Background
Chromosome abnormalities are a characteristic feature of cancer cells. Translocations are large chromosome rearrangements which are key drivers of tumorigenesis and found in various tumor types. Gene fusions result from balanced chromosome translocations and frequently lead to activation and overexpression of an oncogene. The nature of gene fusions strongly correlates with the tumor type, making them very attractive targets for cancer diagnostic or therapeutic intervention.
Chromosomal translocations are triggered in vivo by the simultaneous occurrence of double strand breaks. The scientists at Horizon Discovery have recently published a new robust and precise approach to generating translocations. This advancement facilitates the generation of relevant cell line models for oncology research.
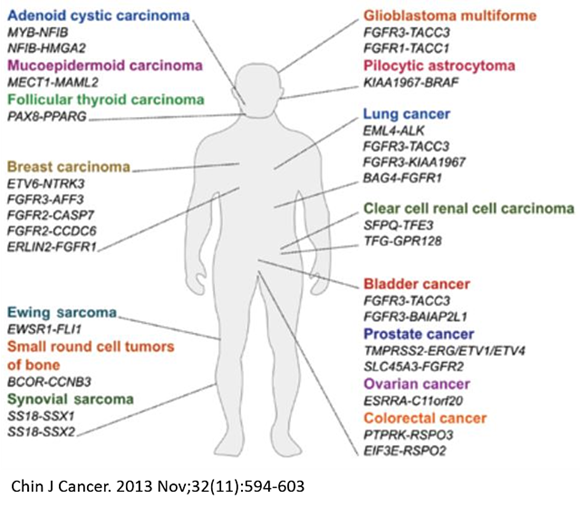
Translocations in Oncology
Creation of de novo cancer relevant translocations
To demonstrate the possibilities of this new approach, we used a human cell line eHAP as a model system to engineer CD74-ROS1 fusion which is frequently found in non-small cell lung cancers. ROS1 is a receptor tyrosine kinase, which is not usually expressed in normal human lung tissue. However, the CD74-ROS1 fusion results the constitutive activation of the ROS1 kinase domain of ROS1, which drives cellular proliferation.
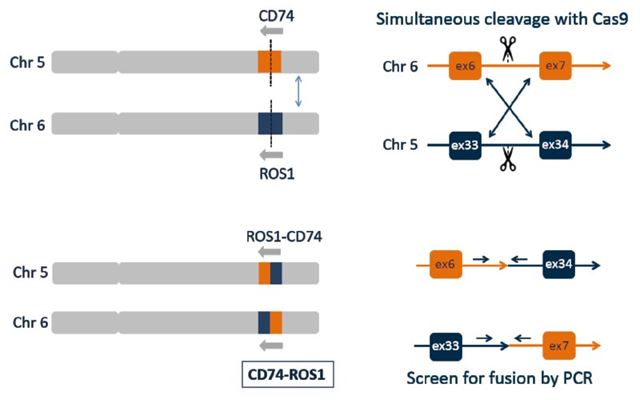
Interchromosomal Translocation Leads to CD74 ROS1 Fusion
To generate such translocations in defined cellular models, we used CRISPR/Cas technology. The method we use is efficient and works with surgical precision both in haploid and diploid backgrounds. By combining two guide RNAs, we introduced two simultaneous double strand breaks that are then spontaneously joined by the endogenous DNA damage repair machinery. Genotyping of hundreds of clones provides a measure for the overall efficiency of the process. Individual clones were characterized in great detail to assess the precision and robustness of the approach we devised.
Correction of an existing translocation
Excitingly, this new technology can also be applied to correcting genetic aberrations. To demonstrate this capability we reverted the BCR-ABL1 fusion (Philadelphia chromosome) back to the wild-type state. BCR-ABL1 fusion (Philadelphia chromosome) was the first cancer specific translocation to be identified. This error involves the balanced translocation between chromosomes 9 and 22. The Philadelphia chromosome is naturally present in eHAP cells, as this is a characteristic feature of chronic myelogenous leukemia, from which the eHAP cell line is derived.
It is conceivable that CRISPR-mediated reversion of an oncogenic fusion event may become a therapeutic strategy in the distant future.
For more information, you can read the full article at BMC Genomics
