Single-gene overexpression experiments have long been considered a perfect complement to information gained in individual loss-of-function studies.
One common strategy is to use an ORF to express an extra copy of the gene of interest that is immune to the loss-of-function effect, also known as a rescue experiment. However, while it seems a logical idea to leverage lentiviral ORF expression approaches for large-scale screening applications, one finds there are a number of unexpected issues with such a plan. Here we provide an explanation of the biggest hurdles to setting up a high-throughput ORF screen, and how an arrayed CRISPRa approach may be a better strategy.
 Equal representation is key in library creation
Equal representation is key in library creation
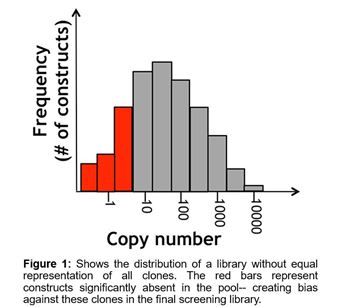
An effective lentiviral screening library must be constructed in a manner where there is a uniform representation of all constructs. This can be especially difficult for lentiviral ORF libraries because the different sized ORF inserts will be packaged with variable efficiency with a tendency to bias towards shorter sequences. If one were to attempt this, then there would need to be a way to calibrate individual constructs within the pools or wells to ensure there is no bias towards or away from certain sequences (as shown in Figure 1). This is a challenge considering the perishable nature of lentivirus, where titer diminishes unpredictably based on time and freeze-thaw cycles. The biggest concerns with using non-uniform material are that there may be a higher false rate of false hits, and less reproducibility between screens. Both of which can result in a great deal of wasted follow-up effort for a researcher.
Beware data deconvolution hurdles
Another point to consider is how to de-convolute the resulting data. If the screening assay is conducted in an arrayed format, then it may straightforward to identify which constructs are associated with individual wells-- as long as a robust phenotypic readout is developed. However, false positives or negatives caused by variations in multiplicity of infection (MOI), should be considered if a lentiviral ORF arrayed platform has variable titer from well-to-well. Past strategies used to control for this variable have involved complex high content image analysis (2), so may not be adaptable to all screening outputs.
If performing the screen in a pooled lentiviral format, next generation sequencing and bioinformatic analysis will be required to identify potential hits. This will require the library-maker to either consider a strategy subcloning ORFs into barcoded vectors for setting up reference FASTA files, or an alternate approach where sequences may be variable in size and sequence and it may be difficult to identify specific ORFs in the population.
The solution: CRISPRa
CRISPRa libraries are an alternative tool that can be used for high throughput increased expression experiments and to avoid the pitfalls mentioned above. The synthetic RNA technology (instead of lentivirus) allows surety of a consistent stock concentration from well-to-well. Delivery into cells is a simple transfection, so uniform performance across a plate and among replicates is better assured. The CRISPRa library system is provided in an arrayed format (one gene/design per well), allowing for easy screen deconvolution, and more complex phenotypic assays than pooled systems (3). Individual lentiviral sgRNA constructs are also available for cells or experiments that require this approach. By expressing a gene directly from the cells native DNA, you can be certain that the isoform is relevant to the cellular biology and it negates challenges associated with promoter function, selection markers, etc. CRISPRa libraries are available for a wide selection of gene families and pathways up to whole genome coverage and custom Cherry-pick libraries can be easily created. Thus also eliminating the time consuming and costly steps of cloning and validating a pooled lentiviral library and speeding your research to critical conclusions and decision points.
Written by Jennifer Abarca PhD, Lead Scientific Support Specialist
Jennifer Abarca has been assisting customers with their questions regarding our product portfolios for over a decade. She believes CRISPRa is one of the most exciting technologies introduced in recent time, "For years we've had researchers requesting an easier way to perform increased gene function screens, and this CRISPRa platform finally delivers a solution that eliminates the difficulties associated with lentiviral and ORF-based methods." When not guiding customers through their experimental journey, Jennifer enjoys building neighborhood camaraderie by acting as director of the local community garden.
Featured Resources
- CRISPRa, cDNA, or ORF? - Blog post
Featured Products
References
- Vermeulen, et al. Pooled shRNA Screening. Frontiers in RNAi, Vol. 1, 2014, 58-78
- Škalamera D., Dahmer-Heath M., Stevenson A. J., Pinto C., Shah E. T., Daignault S. M., Said N. B.M., Davis M., Haass N. K., Williams E. D., Hollier B. G., Thompson E. W., Gabrielli B., et al Genome-wide GOF screen for genes that induce epithelial-to-mesenchymal transition in breast cancer. Oncotarget. 2016; 7: 61000-61020. Retrieved from https://www.oncotarget.com/article/11314/text/
- Žaklina Strezoska, et al. CRISPR-mediated transcriptional activation with synthetic guide RNA. Journal of Biotechnology. Volume 319, 25-35. (2020). https://doi.org/10.1016/j.jbiotec.2020.05.005

