- CRISPR interference reagents
- dCas9-SALL1-SDS3 lentiviral particles
CRISPRmod CRISPRi dCas9-SALL1-SDS3 lentiviral particles
Purified lentiviral particles for generation of stable dCas9-SALL1-SDS3 expressing cell populations

CRISPRi dCas9-SALL1-SDS3 lentiviral particles express a human codon-optimized version of the nuclease-deactivated Cas9 gene, fused to proprietary transcriptional repressors (SALL1 and SDS3). When paired with a well-designed guide RNA that targets a gene near the native transcription start site, expression is repressed.
Review our CRISPRi applications page to get an overview of the technology!
Highlights of using dCas9-SALL1-SDS3 lentiviral particles
- Generate your own stable "CRISPRi ready" dCas9-SALL1-SDS3 expressing cell line.
- Concentrated, purified lentiviral particles are ready for immediate transduction (minimum ≥1×107 TU/mL functional titer, by qPCR).
- Co-expression of blasticidin resistance gene allows for easy cell population selection or for clonal expansion of individual cells.
- Choose from one of three different promoters for optimum expression in your cells of interest (see options below).
Requirements for a CRISPRi experiment using dCas9-SALL1-SDS3 lentiviral particles
- CRISPRi dCas9-SALL1-SDS3 lentiviral particles
- CRISPRi lentiviral sgRNA for your target gene (see Workflow tab)
Not all RNA pol II promoters are equally active in different cellular environments
The activity of any given promoter controlling the transcription of dCas9-SALL1-SDS3 can differ greatly from one biological context to another, resulting in variable dCas9-SALL1-SDS3 expression levels and thus varying levels of gene repression. Choosing an optimal promoter for your cell line will be important for robust gene overexpression.
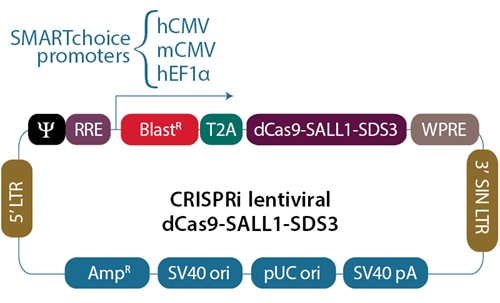
SMARTchoice promoter options for expressing dCas9-SALL1-SDS3
| Promoter | Description |
|---|---|
| hCMV | human cytomegalovirus immediate early promoter |
| mCMV | mouse cytomegalovirus immediate early promoter |
| hEF1α | human elongation factor 1 alpha promoter |
CRISPRi workflow using synthetic sgRNA and dCas9-SALL1-SDS3 lentiviral particles
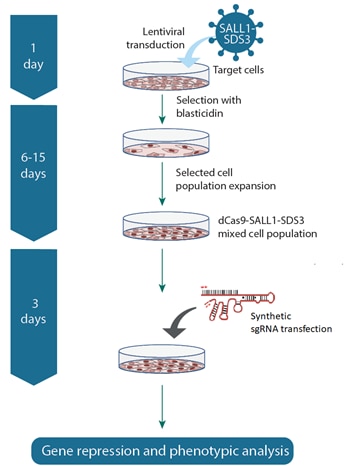
Transduce cells with CRISPRi dCas9-SALL1-SDS3 lentiviral particles to generate CRISPRi-ready, stable dCas9-SALL1-SDS3 expressing cells. Then introduce CRISPRi synthetic sgRNA specific to your target gene(s). This system is ideal for arrayed screening in transfectable cell models.
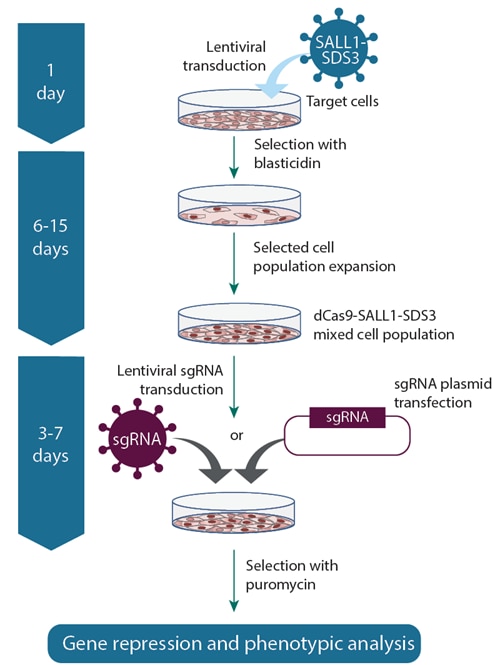
CRISPRi workflow using lentiviral sgRNA and dCas9-SALL1-SDS3 lentiviral particles
Workflow using CRISPRi dCas9-SALL1-SDS3 lentiviral particles to establish a stable dCas9-SALL1-SDS3 expressing cell line, then delivering CRISPRi lentiviral sgRNA particles (left) or plasmid sgRNA (right). This system is ideal for working in difficult-to-transfect cell types.
CRISPRi with dCas9-SALL1-SDS3 results in more robust protein level repression than CRISPRi with dCas9-KRAB
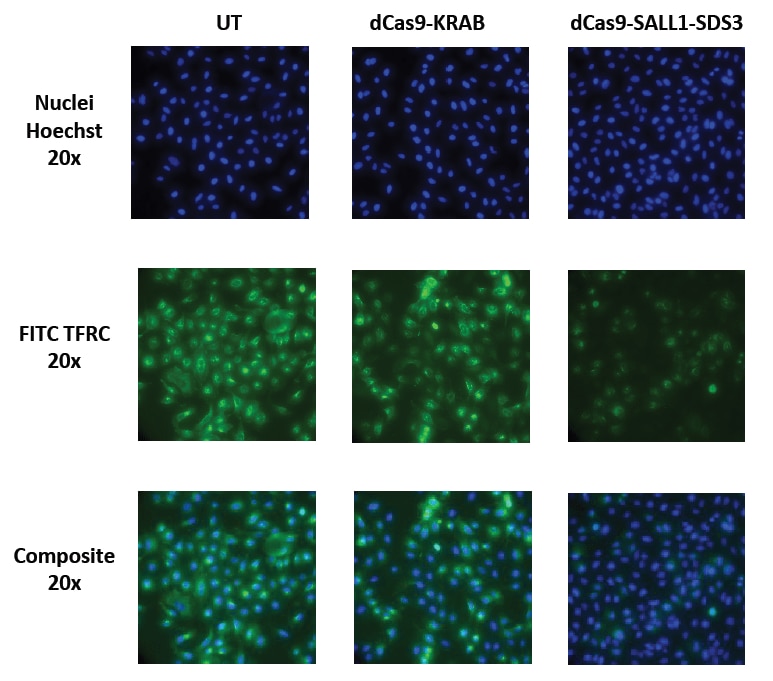
Functional knockdown of TFRC was assessed in U2OS cells constitutively expressing either dCas9-KRAB or dCas9-SALL1-SDS3 under the hEF1α promoter. Cells were transfected with a 25 nM pool containing three CRISPRi synthetic sgRNAs targeting TFRC using DharmaFECT 4 Transfection Reagent. 72 hours post-transfection the cells were split. 96 hours post-transfection the cells were fixed and stained with a primary antibody targeting TFRC and an Alexa Fluor 488 conjugated fluorescent secondary antibody. Hoechst stain was used to identify nuclei.
Cells expressing dCas9-SALL1-SDS3 demonstrate greater and more consistent CRISPRi gene repression compared to dCas9-KRAB
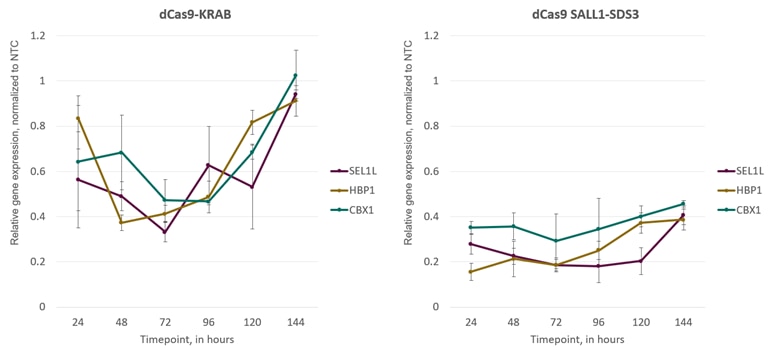
CRISPRi synthetic sgRNAs achieve maximal repression at 48-72 hours post-transfection in both dCas9-KRAB and dCas9-SALL1-SDS3-expressing cells. U2OS cells stably expressing integrated dCas9-KRAB or dCas9-SALL1-SDS3 were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with pools of CRISPRi synthetic sgRNA (25nM) targeting CBX1, HBP1, or SEL1L. Cells were harvested at 24, 48, 72, 96, 120, and 144 hours post-transfection, total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative expression of each target gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).
LentiBOOST Lentivirus Transduction Enhancer is a uniquely formulated transduction reagent that can be used with or without lentivirus spinfection in order to increase successful viral transduction events while preserving cell viability. Especially critical for preserving precious primary cells from patient cohorts, or, for engineering complex animal models; improving transduction efficiency can save time and costs by increasing the success of each editing/transduction step, or, even avoid the loss of irreplaceable samples. Additionally, LentiBOOST technology is already used in the manufacturing of a number of clinical stage therapies providing the opportunity to demonstrate improved workflow applicability to the clinic.
LentiBOOST can be purchased through the Dharmacon Reagents catalog.
To learn more about LentiBOOST technology visit the Revvity LentiBOOST webpage.
Supporting Data
Improved CD8+ T-cell SMARTvector™ shRNA lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
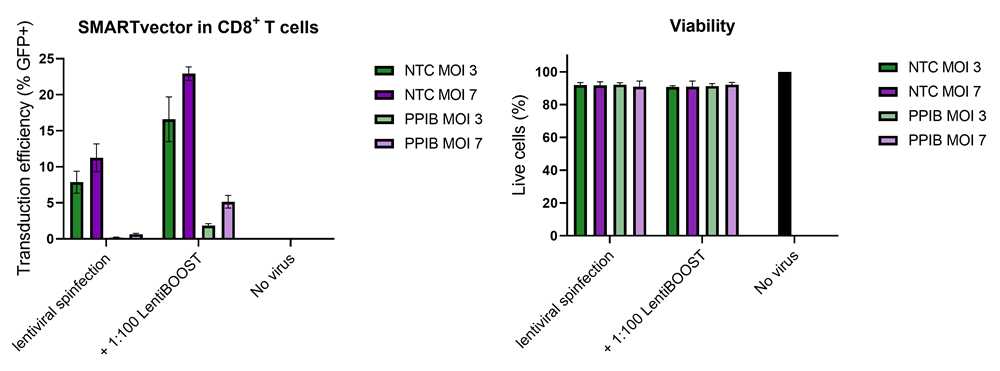
100,000 primary human CD8+ T cells were transduced with either 30,000 (MOI 3, green) or 70,000 (MOI 7, purple) TUs of SMARTVector™ mCMV tGFP Lentiviral Control Particles targeting either NTC or PPIB along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by a four hour incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency (%GFP+ out of live cells) and viability were determined 5 days post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved CD4+ and CD8+ T-cell Edit-R™ All-in-one sgRNA/Cas9 lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
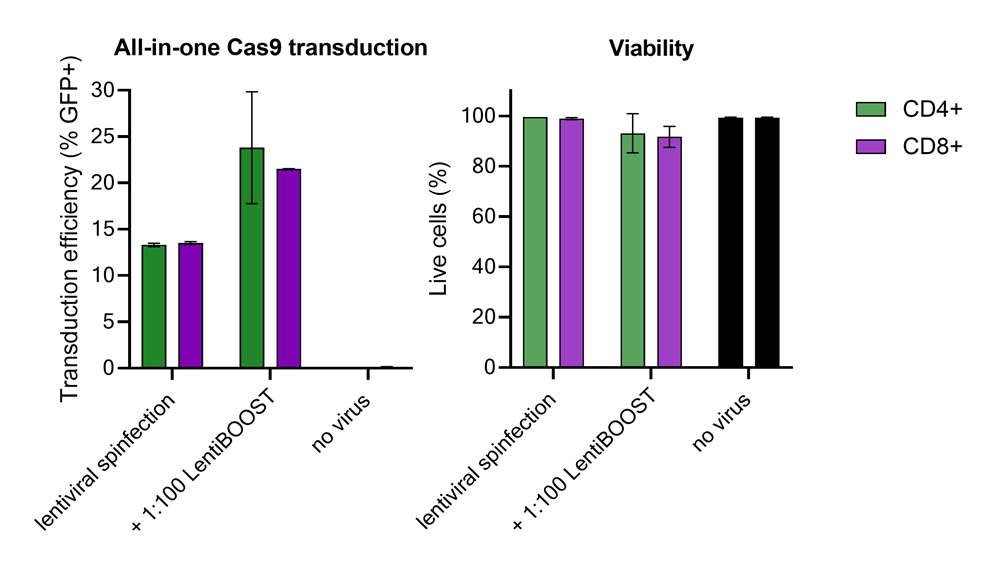
100,000 primary human CD4+ and CD8+ T cells from two donors were transduced with 250,000 TUs of Edit-R GFP Delivery controls mCMV along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved transduction of human induced pluripotent stem cells (hiPSCs) with the Strict-R™ Inducible CRISPRa lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer

10,000 WTC hiPS cells were transduced with either 20,000 (MOI 2, green) or 40,000 (MOI 4, purple) TUs of Strict-R™ Inducible EGFP dCas9-VPR Lentiviral Particles along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST markedly improved transduction efficiencies without significantly impacting cell viability.
Safety data sheets
Selection guides
Related Products
Predesigned CRISPRi lentiviral sgRNAs for highly efficient gene repression of any human gene.
Validated CRISPRi lentiviral sgRNA for optimization of transcriptional repression experiments.
Validated CRISPRi lentiviral sgRNA for evaluation of transcriptional repression experiments.
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.