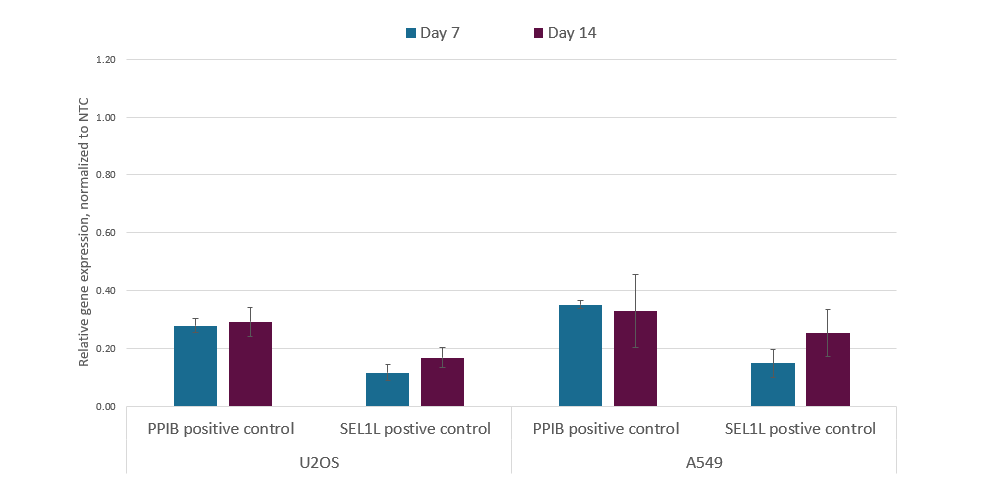- CRISPR interference reagents
- CRISPRi lentiviral sgRNA
CRISPRmod CRISPRi lentiviral sgRNA
Predesigned CRISPRi lentiviral sgRNAs for highly efficient gene repression of any human gene
- An ideal delivery method for CRISPRi in difficult-to-transfect cells, or when longer term repression is desired
- Available as high-titer lentiviral particles or glycerol stock

CRISPRi lentiviral sgRNA
1Start Here
2Choose
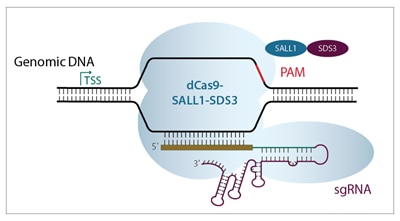
Efficient endogenous gene repression with CRISPRi
CRISPR interference (CRISPRi) is a unique adaptation of the CRISPR-Cas9 gene editing system. The Horizon CRISPRmod CRISPRi system utilizes a catalytically deactivated Cas9 (dCas9) fused to our proprietary transcriptional repressors (SALL1 and SDS3). When paired with an algorithm-designed guide RNA that targets a gene immediately downstream of the transcriptional start site (TSS), it promotes transcriptional repression.
Review our CRISPRi applications page to get an overview of the technology!
Highlights of CRISPRi lentiviral sgRNA reagents
- Available as glycerol stocks or high-titer purified lentivirus particles
- Designed with a published algorithm by Horlbeck, et. al. (2016) that has demonstrated strong levels of gene repression from optimized designs
- Purified, concentrated high-titer lentivirus particles can be directly transduced into cells, including difficult-to-transfect cells and lines
Requirements for a CRISPRi experiment using lentiviral sgRNA
- CRISPRi lentiviral sgRNA for your target gene
- CRISPRi dCas9-SALL1-SDS3 lentiviral particles (see Workflow tab)
Schematic diagram of the CRISPRi lentiviral sgRNA vectors
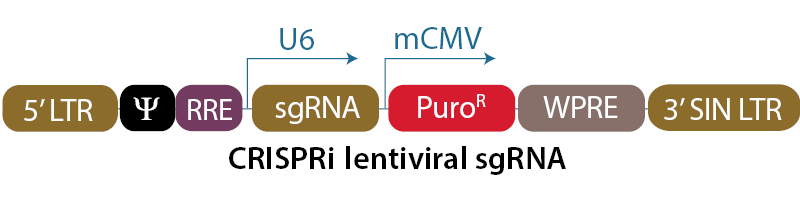
In the CRISPRi lentiviral sgRNA vector backbone, the gene-specific guide RNA is expressed under the control of a human U6 promoter, while expression of the puromycin resistance gene (PuroR) is driven from the mouse CMV promoter and allows for rapid selection of cells with integrated sgRNA. The plasmid contains the AmpR resistance marker for growth and selection in E. coli.
CRISPRi lentiviral sgRNA workflow
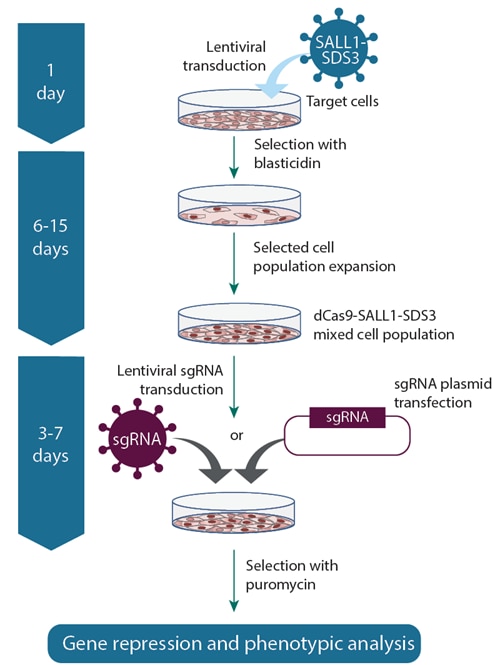
Workflow using CRISPRi dCas9-SALL1-SDS3 lentiviral particles to establish a stable dCas9-SALL1-SDS3 expressing cell line, then delivering CRISPRi lentiviral sgRNA particles (left) or plasmid sgRNA (right).
CRISPRi lentiviral sgRNA controls
CRISPRi lentiviral sgRNA positive controls
- CRISPRi sgRNAs targeting well-characterized genes to determine the effectiveness of your experimental conditions for maximum repression.
CRISPRi lentiviral sgRNA non-targeting controls
- CRISPRi non-targeting controls to evaluate baseline cellular responses to CRISPRi components in the absence of target-specific sgRNA.
Related products
CRISPRi dCas9-SALL1-SDS3
- Lentiviral particles or mRNA that express nuclease-deactivated Cas9 fused to our proprietary transcriptional repressors for CRISPRi experiments.
CRISPRi lentiviral sgRNA is well suited for long term repression
U2OS and A549 cells stably expressing integrated dCas9-KRAB or dCas9-SALL1-SDS3 were plated at 50,000 cells/well in 24 well plates and transduced with CRISPRi sgRNA expressing lentiviral particles targeting PPIB or SEL1L at a MOI of 0.3 to obtain cells with a single integrant. Cells were selected with 2 µg/mL puromycin for 5 days, split into two populations, and allowed to recover for an additional day. At 7 days post-transduction one population was harvested for RT-qPCR analysis. The replicate populations of cells were cultured for 7 additional days before harvest. Total RNA was isolated from harvested plates and relative gene expression was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
- M. A. Horlbeck et al., Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760 (2016).
LentiBOOST Lentivirus Transduction Enhancer is a uniquely formulated transduction reagent that can be used with or without lentivirus spinfection in order to increase successful viral transduction events while preserving cell viability. Especially critical for preserving precious primary cells from patient cohorts, or, for engineering complex animal models; improving transduction efficiency can save time and costs by increasing the success of each editing/transduction step, or, even avoid the loss of irreplaceable samples. Additionally, LentiBOOST technology is already used in the manufacturing of a number of clinical stage therapies providing the opportunity to demonstrate improved workflow applicability to the clinic.
LentiBOOST can be purchased through the Dharmacon Reagents catalog.
To learn more about LentiBOOST technology visit the Revvity LentiBOOST webpage.
Supporting Data
Improved CD8+ T-cell SMARTvector™ shRNA lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
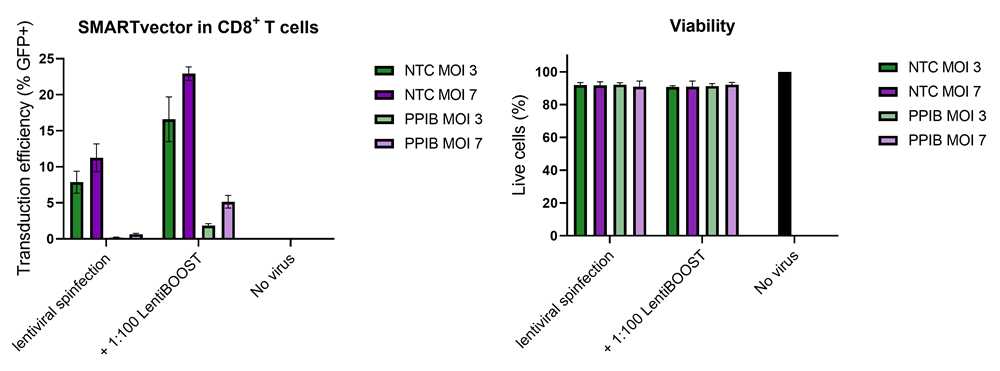
100,000 primary human CD8+ T cells were transduced with either 30,000 (MOI 3, green) or 70,000 (MOI 7, purple) TUs of SMARTVector™ mCMV tGFP Lentiviral Control Particles targeting either NTC or PPIB along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by a four hour incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency (%GFP+ out of live cells) and viability were determined 5 days post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved CD4+ and CD8+ T-cell Edit-R™ All-in-one sgRNA/Cas9 lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
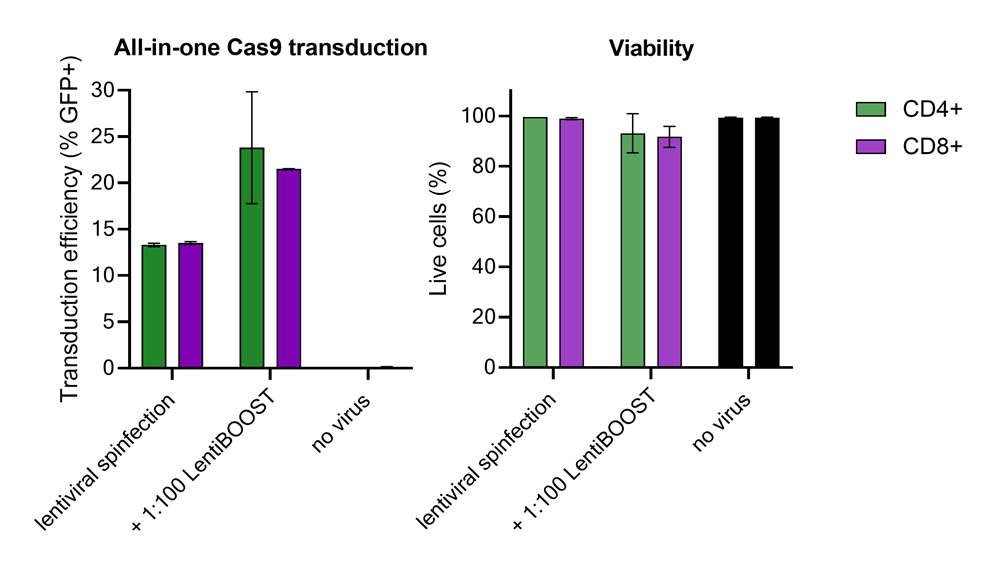
100,000 primary human CD4+ and CD8+ T cells from two donors were transduced with 250,000 TUs of Edit-R GFP Delivery controls mCMV along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved transduction of human induced pluripotent stem cells (hiPSCs) with the Strict-R™ Inducible CRISPRa lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer

10,000 WTC hiPS cells were transduced with either 20,000 (MOI 2, green) or 40,000 (MOI 4, purple) TUs of Strict-R™ Inducible EGFP dCas9-VPR Lentiviral Particles along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST markedly improved transduction efficiencies without significantly impacting cell viability.
Product inserts
Safety data sheets
Technical manuals
Related Products
Validated CRISPRi lentiviral sgRNA for optimization of transcriptional repression experiments.
Validated CRISPRi lentiviral sgRNA for evaluation of transcriptional repression experiments.
Purified lentiviral particles for generation of stable dCas9-SALL1-SDS3 expressing cell populations.
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.