- CRISPR interference reagents
- CRISPRi synthetic sgRNA
CRISPRmod CRISPRi synthetic sgRNA
Predesigned CRISPRi guide RNAs for highly specific gene repression of any human protein-coding gene
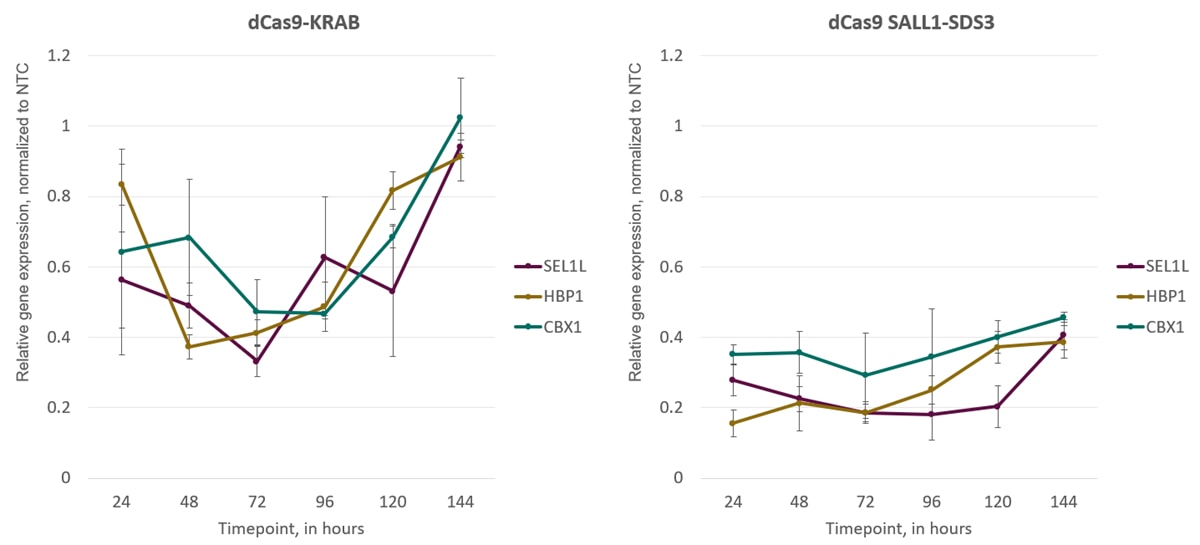
- Our CRISPRi system has been developed to outperform other systems (KRAB and MECP2) when combined with synthetic guides - observe repression within 24 hours post-transfection
- Co-transfect or electroporate with CRISPRi dCas9-SALL1-SDS3 mRNA for a lentiviral-free CRISPRi workflow
- Available as pooled or individual reagents

CRISPRi synthetic sgRNA
1Start Here
2Choose
Efficient transcriptional gene repression with CRISPRi
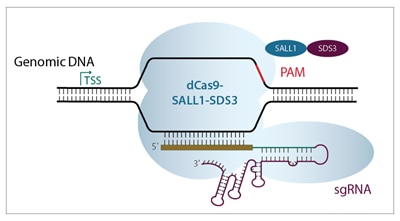
CRISPR interference (CRISPRi) is a unique adaptation of the CRISPR-Cas9 gene editing system. The Horizon CRISPRmod CRISPRi system utilizes a catalytically deactivated Cas9 (dCas9) fused to our proprietary transcriptional repressors (SALL1 and SDS3). When paired with an algorithm-designed guide RNA that targets a gene immediately downstream of the transcriptional start site (TSS), it promotes transcriptional repression.
Review our CRISPRi applications page to see an overview of the technology!
Highlights of CRISPRi synthetic sgRNA reagents
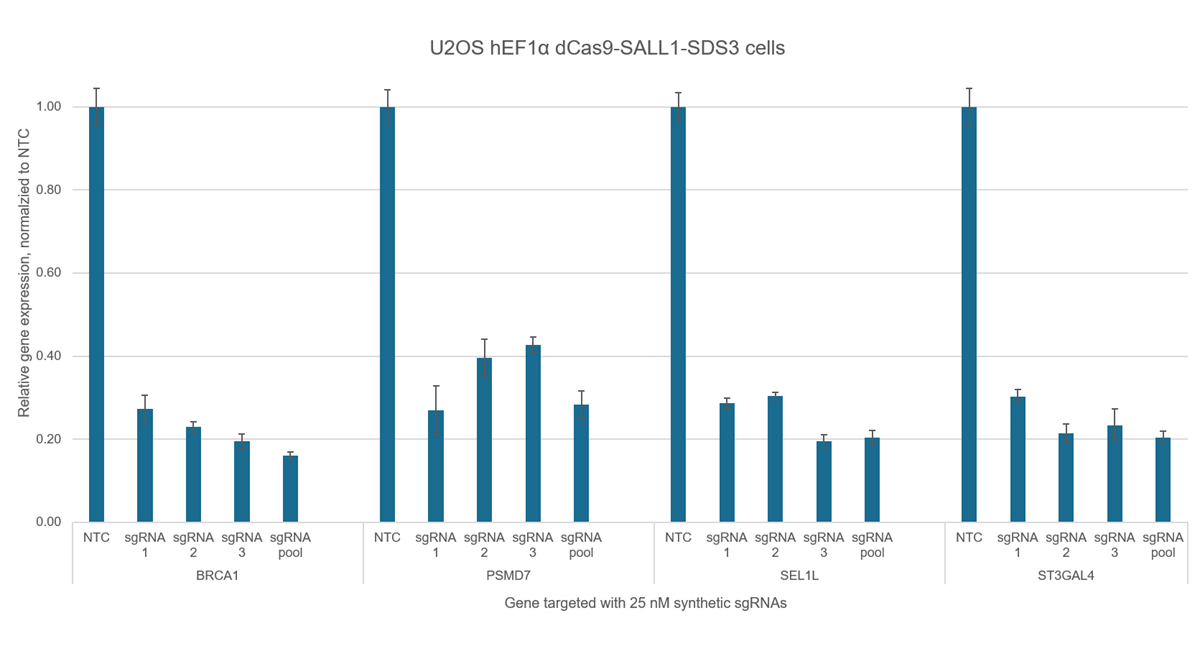
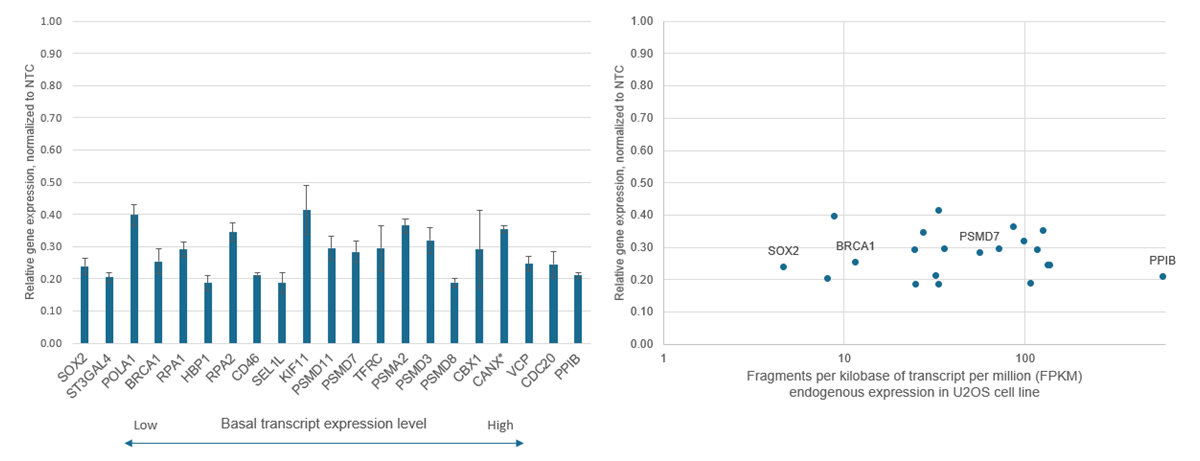
- CRISPR-based PAM-anchored targeting provides highly specific transcriptional repression (see Supporting data tab)
- Designs optimized from a published algorithm by Horlbeck, et. al. demonstrate strong levels of gene repression (see References tab)
- Chemically modified CRISPRi sgRNAs provide additional stability against nuclease degradation and improve overall performance
- For genes with alternative characterized start sites, distinct guide RNA designs are available (labeled P2)
Requirements for a CRISPRi experiment using synthetic sgRNA
- CRISPRi synthetic sgRNA for your target gene - choose from individual guides, sets of 3 guides, or pools of 3 guides
- CRISPRi dCas9-SALL1-SDS3 source - choose from mRNA or lentiviral particles (see Workflow tab)
- Appropriate positive and non-targeting controls
- DharmaFECT transfection reagent or electroporation for guide delivery
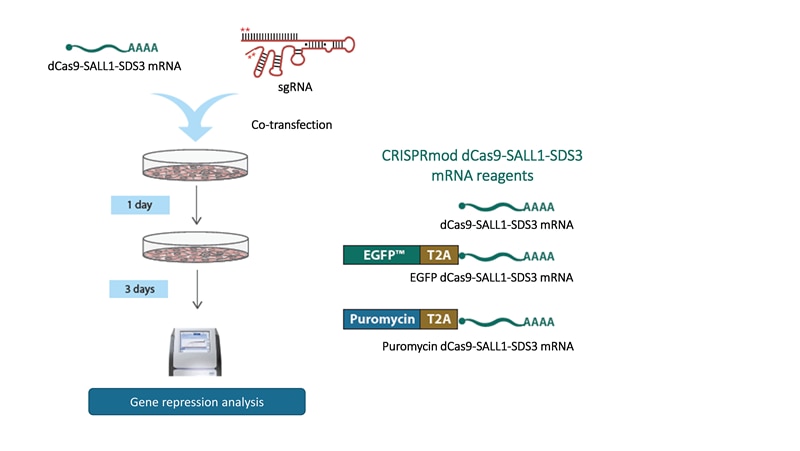
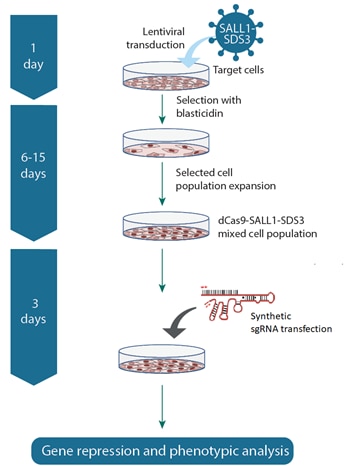
Complete your CRISPRi experiment by adding these supporting reagents to your order
CRISPRi dCas9-SALL1-SDS3
- CRISPRi dCas9-SALL1-SDS3 mRNA can be co-transfected with CRISPRi synthetic sgRNA for a fully DNA-free workflow.
CRISPRi synthetic guide RNA controls
- CRISPRi synthetic sgRNA non-targeting controls are used to evaluate baseline cellular responses to CRISPRi components in the absence of target-specific sgRNA. Choose from 5 scrambled sequences guaranteed not to target any gene in the human genome.
- CRISPRi synthetic sgRNA positive controls targeting well-characterized genes are used to determine the effectiveness of your experimental conditions for maximum repression. Choose from PPIB or SEL1L1.
Transfection reagents
- DharmaFECT transfection reagents are optimized for improved delivery and reduced toxicity. The optimal DharmaFECT reagent for your experiment will depend on your Cas9 nuclease source and your cell type.
| dCas9-SALL1-SDS3 source | Recommended transfection reagent |
|---|---|
| dCas9-SALL1-SDS3 lentiviral particles | DharmaFECT 1,2,3, or 4 for transfection of synthetic guide RNA. Use this guide to find the recommended DharmaFECT reagent for your cell type. |
| dCas9-SALL1-SDS3 mRNA | DharmaFECT Duo for co-transfection of mRNA or protein and synthetic guide RNA |
- M. A. Horlbeck et al., Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760 (2016). https://doi.org/10.7554/eLife.19760
- C. Mills et al., A Novel CRISPR interference effector enabling functional gene characterization with synthetic guide RNAs. The CRISPR Journal. Vol 5, [6] (2022). http://doi.org/10.1089/crispr.2022.0056
Protocols
Quick protocols
Safety data sheets
Technical manuals
Related Products
Validated CRISPRi synthetic pools or individual sgRNA for optimization of transcriptional repression experiments.
Validated CRISPRi synthetic pools or individual sgRNA for evaluation of transcriptional repression experiments.