- Gene editing
- Gene editing reagents
- Lentiviral Cas9 nuclease reagents
Lentiviral Cas9 nuclease reagents
Lentiviral CRISPR-Cas9 components for robust gene editing in biologically relevant cell types
Purified lentiviral particles for generation of stable Cas9 nuclease-expressing cell populations. Inquire for pricing on larger volumes.
Try our new: Dharmacon™ Strict-R™ Inducible Cas9 Lentiviral System for dual controlled gene knockout in diverse cell types.

Cas9 nuclease in the CRISPR-Cas9 system
The CRISPR-associated enzyme Cas9 is an RNA-guided endonuclease that requires a guide RNA for genomic DNA target recognition and generation of DNA double-strand breaks (DSB).
Lentiviral Cas9 reagents facilitate rapid generation of cell lines that express Cas9 nuclease, which empower many gene editing applications, including: pooled lentiviral sgRNA screening, arrayed screening with crRNA libraries, or assessment of multiple guide RNAs targeting a single gene and/or multiple genes.
The lentiviral Cas9 nuclease expression vectors contain a human codon-optimized version of S. pyogenes Cas9 nuclease under the control of various constitutive, or an inducible promoter. All formats can be pre-packaged as purified lentiviral particles.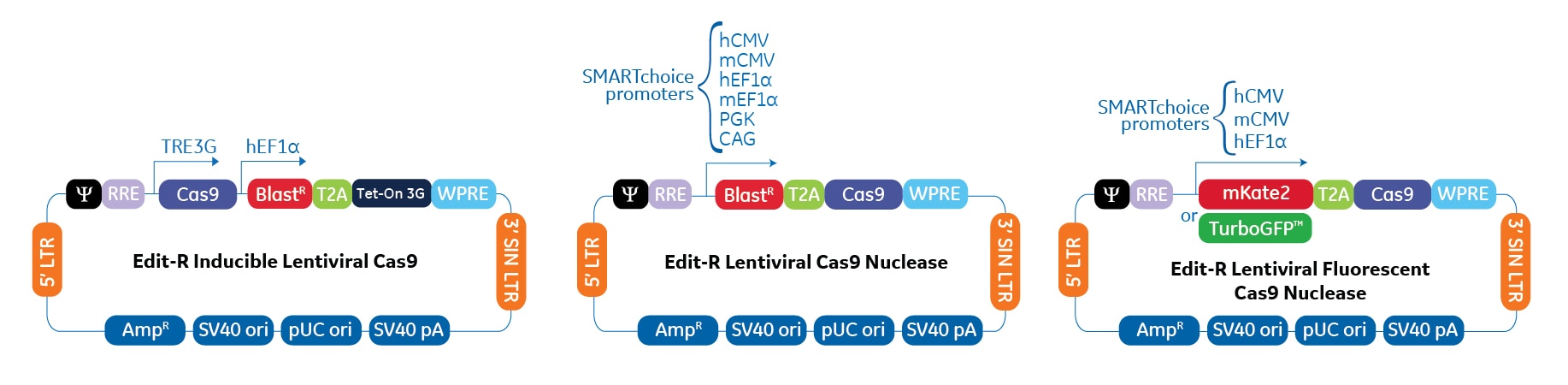
Lentiviral Cas9 expression vector highlights
- Selection using blasticidin resistance marker (BlastR) or fluorescent marker (mKate2 or TurboGFP™)
- Provided as concentrated, purified lentiviral particles for immediate transduction, with a minimum ≥ 1 × 107 TU/mL functional titer.
- Customize your construct with one of six SMARTchoice constitutive promoters to ensure optimal Cas9 expression in your cell line of interest
- Utilize tight regulation of the inducible Cas9 vector when you require temporal control over the expression of Cas9, or to create a stable cell line with minimal background expression.
Not all RNA pol II promoters are equally active in different cellular environments
The activity of any given promoter controlling the transcription of Cas9 nuclease can differ greatly from one biological system to another, resulting in variable Cas9 expression levels and thus varying levels of DNA editing. Choosing an optimal promoter for your cell line or type will therefore affect the degree of gene editing in your experiment.
SMARTchoice promoter options for expressing Cas9 nuclease| Promoter | Description |
|---|---|
| hCMV | human cytomegalovirus immediate early promoter |
| mCMV | mouse cytomegalovirus immediate early promoter |
| hEF1α | human elongation factor 1 alpha promoter |
| mEF1α | mouse elongation factor 1 alpha promoter |
| PGK | mouse phosphoglycerate kinase promoter |
| CAG | chicken beta actin hybrid promoter |
| TRE3G | doxycycline-inducible promoter |
Gene editing workflows for lentiviral Cas9 nuclease reagent formats
Gene knockout workflow using the lentiviral Cas9 nuclease with Edit-R lentiviral sgRNA
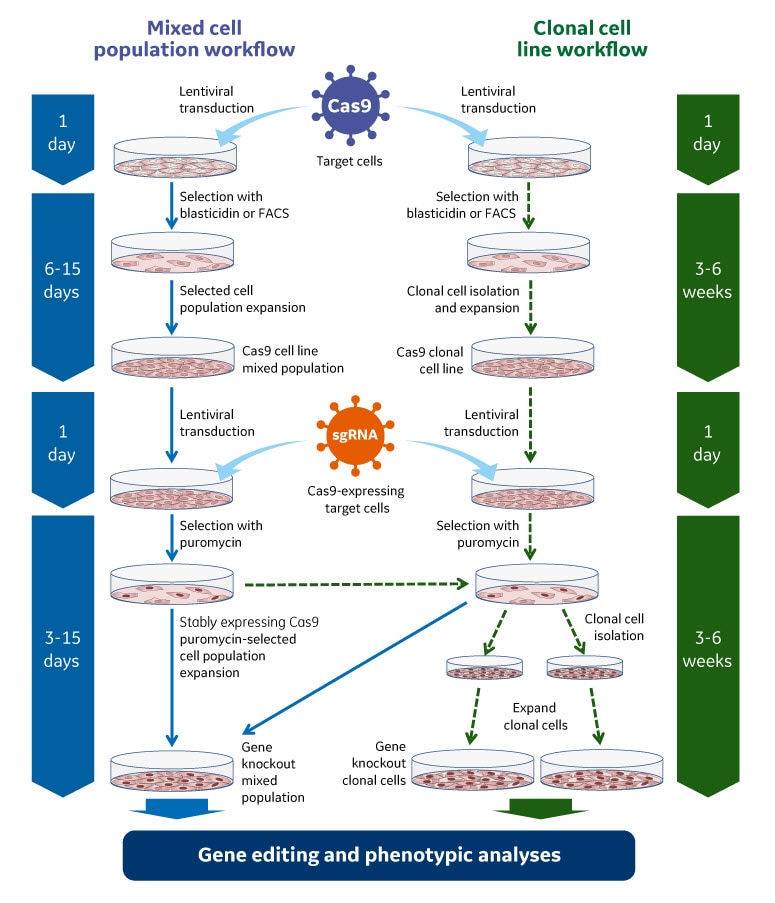
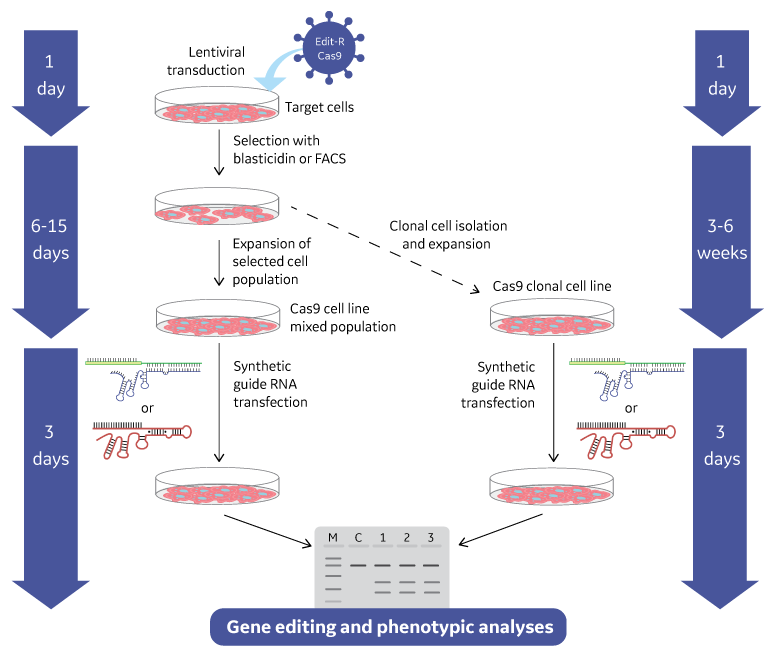
Gene knockout workflow using the lentiviral Cas9 nuclease with sgRNA system. Gene editing can be performed using a mixed cell population approach (left side) typically for gene knockout screening or an isolated clonal cell line approach (right side) when a more defined cell type is required for phenotypic analysis.
Gene knockout workflow using the inducible lentiviral Cas9 nuclease with Edit-R lentiviral sgRNA
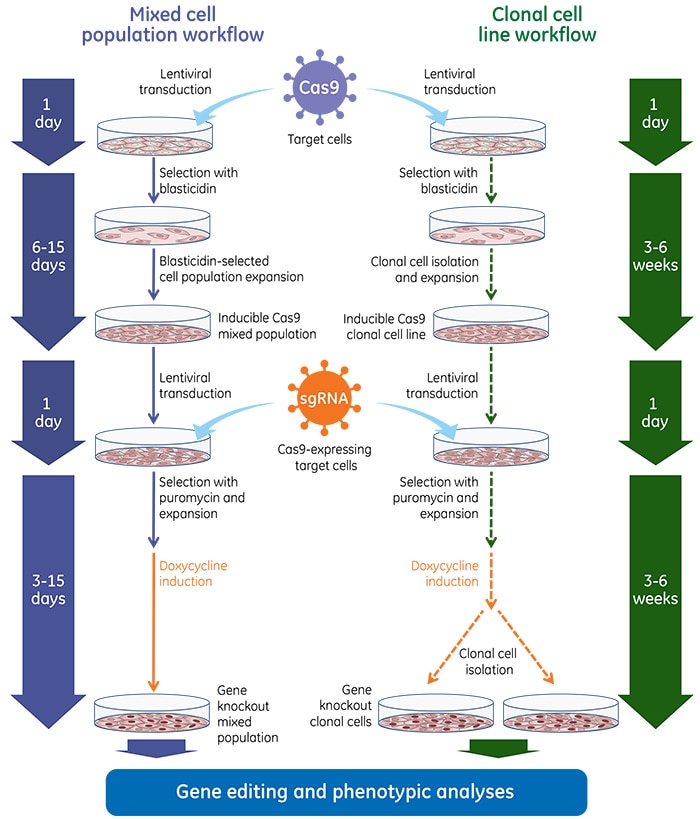
Gene knockout workflow using the lentiviral Cas9 nuclease with Edit-R synthetic guide RNA
Gene knockout workflow using the inducible lentiviral Cas9 nuclease in a pooled lentiviral sgRNA screen
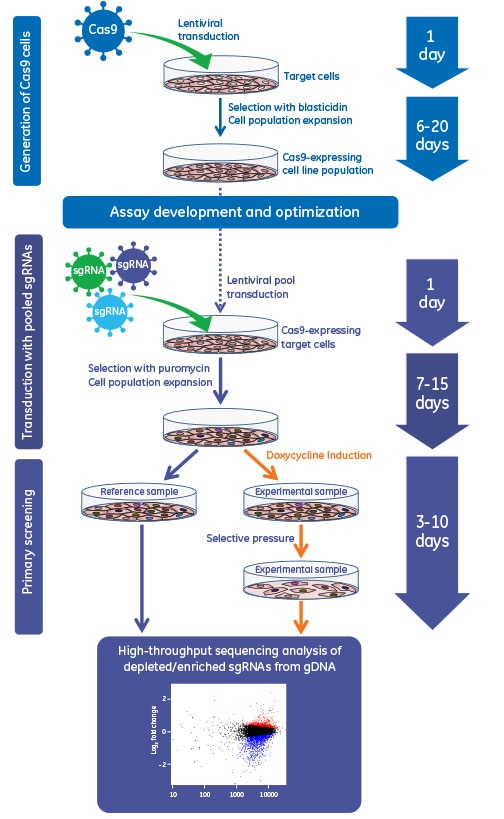
Lentiviral fluorescent Cas9 nuclease data
Fluorescent lentiviral Cas9 nuclease enables enrichment for high expression and improved CRISPR-Cas9 gene editing efficiency
U2OS cells were transduced at low multiplicity of infection (MOI 0.3) with Edit-R Lentiviral hCMV mKate2-Cas9 Nuclease particles (Cat #VCAS11869) so that transduced cells would have only one integration of Cas9. Cells were expanded for fluorescence activated cell sorting (FACS) where populations were sorted into negative, low, medium and high mKate2 fluorescence. These subpopulations were expanded and then plated at 10,000 cells/well in a 96-well plate. One day later, cells were transfected with tracrRNA (25 nM, Cat #U-002005-xx) and Edit-R PPIB Synthetic crRNA Control (Cat #U-007501-xx) or Edit-R MYC Predesign crRNA (Cat #CM-003282-01) using DharmaFECT 1 transfection reagent (0.3 µL/well, Cat #T-2001-01). After 72 hours, cells were imaged for mKate2 fluorescence using the In Cell Analyzer 2200 (GE Healthcare; A) and then harvested for DNA mismatch detection assay to estimate gene editing (B). High mKate2 expression can be associated with the highest levels of gene editing for both PPIB- and MYC-targeting crRNAs.
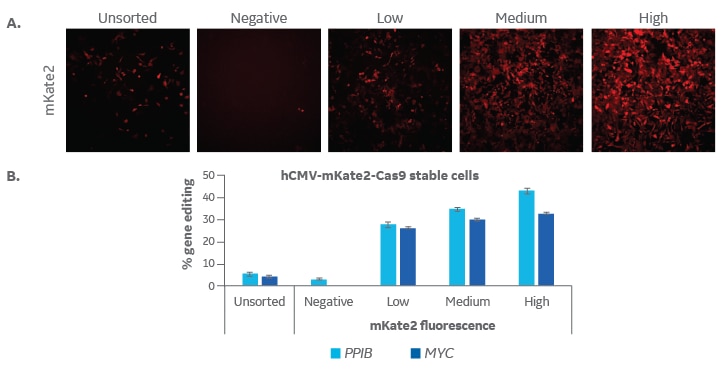
Lentiviral inducible Cas9 nuclease data
Gene editing activity of inducible Cas9 vectors after induction with Doxycycline for 7 days
Cells were transduced with a constitutive (CAG-Cas9) or an inducible (TRE3G-Cas9) Cas9 expression lentiviral particles at an MOI of 0.3, and selected with 10 µg/mL blasticidin in tetracycline-free medium for 10 days. Cas9-stable cells were then transduced with DNMT3B- or PPIB-sgRNA lentiviral particles at an MOI of 0.3. Cells were selected with 2 µg/mL puromycin for 2 days in tetracycline-free medium and split in two populations: uninduced (DOX-) and induced (DOX+) with 500 ng/mL doxycycline for 7 days. The cells were then lysed and analyzed for indels using a DNA mismatch detection assay with T7EI.
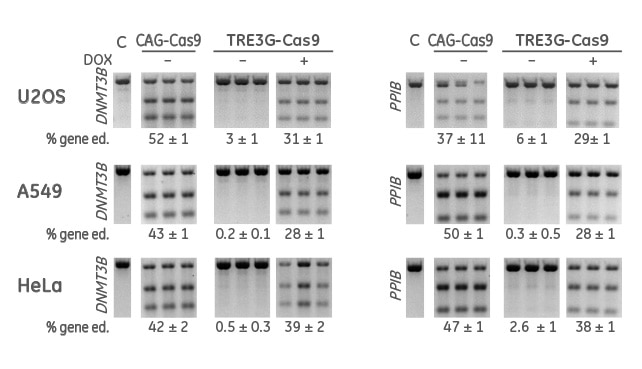
Inducible Cas9 nuclease vector displays minimal leak after 21 days without doxycycline induction
Cells were transduced with a constitutive (CAG-Cas9) or an inducible (TRE3G-Cas9) Cas9 expression lentiviral particles at an MOI of 0.3, and selected with 10 µg/mL blasticidin in tetracycline-free medium for 10 days. Cas9-stable cells were then transduced with DNMT3B- or PPIB-sgRNA lentiviral particles at an MOI of 0.3. Cells were selected with 2 µg/mL puromycin and maintained in tetracycline-free medium for 21 days. The cells were then lysed and analyzed for indels using a DNA mismatch detection assay with T7EI.
Dose response for doxycycline in inducible U2OS-Cas9 cells
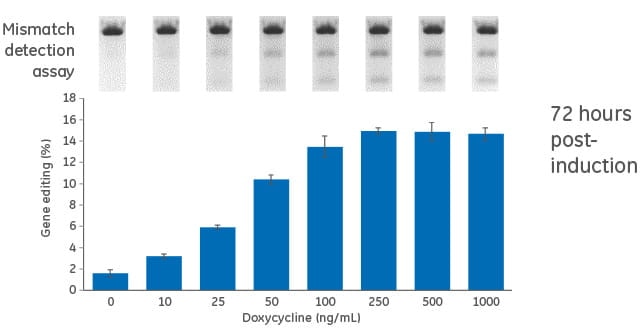
Cells were transduced with the inducible (TRE3G-Cas9) Cas9 expression lentiviral particles at an MOI of 0.3, and selected with 10 µg/mL blasticidin in tetracycline-free medium for 10 days. Cas9-stable cells were then transduced with PPIB-sgRNA lentiviral particles at an MOI of 0.3. Cells were selected with 2 µg/mL puromycin tetracycline-free medium for 4 days, suspended with trypsin and seeded in a 96-well plate in medium containing increasing concentrations of doxycycline (0 to 1000 ng/mL). The cells were incubated for 72 hours, lysed and analyzed for indel formation using a DNA mismatch detection assay with T7EI. Upper panel, representative gel image of the DNA mismatch detection assay with T7EI for PPIB targeted amplicon; lower panel, mean ± standard deviation of the estimated percentage of gene editing from three independently treated wells.
Lentiviral Cas9 nuclease data
Differential expression of Cas9 by different promoters induce varying levels of gene editing
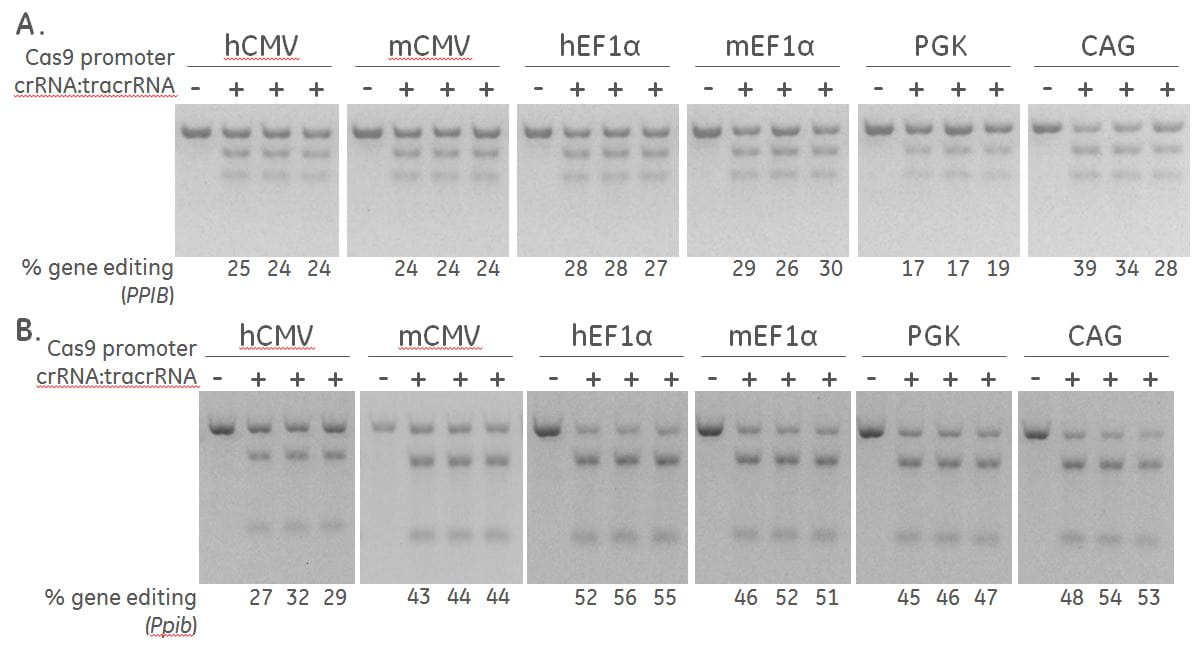
A human recombinant U2OS ubiquitin-EGFP proteasome cell line (Ubi[G76V]-EGFP) (A) and a mouse fibroblast (NIH/3T3) (B), were stably transduced with lentiviral particles containing Cas9 and a blasticidin resistance gene driven by the indicated promoters.. A population of cells with stably integrated Cas9-blastR was selected with blasticidin for a minimum of 10 days before transfections. Cells were transfected with 50 nM synthetic crRNA:tracrRNA targeting Human PPIB / mouse Ppib using DharmaFECT 1 and DharmaFECT 3 Transfection reagent, respectively. After 72 hours, the relative frequency of gene editing was calculated based on a DNA mismatch detection assay using T7EI on genomic DNA extracted from the transfected cells.
- D. Bhaya, et al. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet.45, 273-297 (2011).
- M. Jinek, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science337, 816-821 (2012).
- E. Deltcheva, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor Nuclease III. Nature471, 602-607 (2011).
- P. Mali, et al. RNA-guided human genome engineering via Cas9. Science339, 823-826 (2013).
- Y. Fu, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol.31, 822-826 (2013).
- P.D. Hsu, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol.31, 827-832 (2013).
- T. Wang et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science343, 80-84 (2014).
- D.Y. Guschin, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol.649, 247-256 (2010).
- L.Cong, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science339, 819-823 (2013).
- J.C. Kappes, X. Wu and J.K. Wakefield. Production of trans-lentiviral vector with predictable safety. Methods Mol. Med.76, 449-465 (2003).
- R.H. Kutner, X.-Y. Zhang and J. Reiser. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc.4, 495-505 (2009).
LentiBOOST Lentivirus Transduction Enhancer is a uniquely formulated transduction reagent that can be used with or without lentivirus spinfection in order to increase successful viral transduction events while preserving cell viability. Especially critical for preserving precious primary cells from patient cohorts, or, for engineering complex animal models; improving transduction efficiency can save time and costs by increasing the success of each editing/transduction step, or, even avoid the loss of irreplaceable samples. Additionally, LentiBOOST technology is already used in the manufacturing of a number of clinical stage therapies providing the opportunity to demonstrate improved workflow applicability to the clinic.
LentiBOOST can be purchased through the Dharmacon Reagents catalog.
To learn more about LentiBOOST technology visit the Revvity LentiBOOST webpage.
Supporting Data
Improved CD8+ T-cell SMARTvector™ shRNA lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
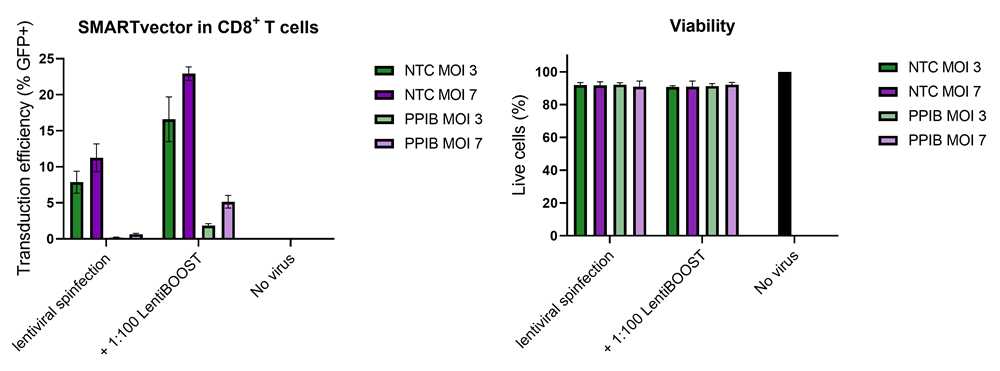
100,000 primary human CD8+ T cells were transduced with either 30,000 (MOI 3, green) or 70,000 (MOI 7, purple) TUs of SMARTVector™ mCMV tGFP Lentiviral Control Particles targeting either NTC or PPIB along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by a four hour incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency (%GFP+ out of live cells) and viability were determined 5 days post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved CD4+ and CD8+ T-cell Edit-R™ All-in-one sgRNA/Cas9 lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
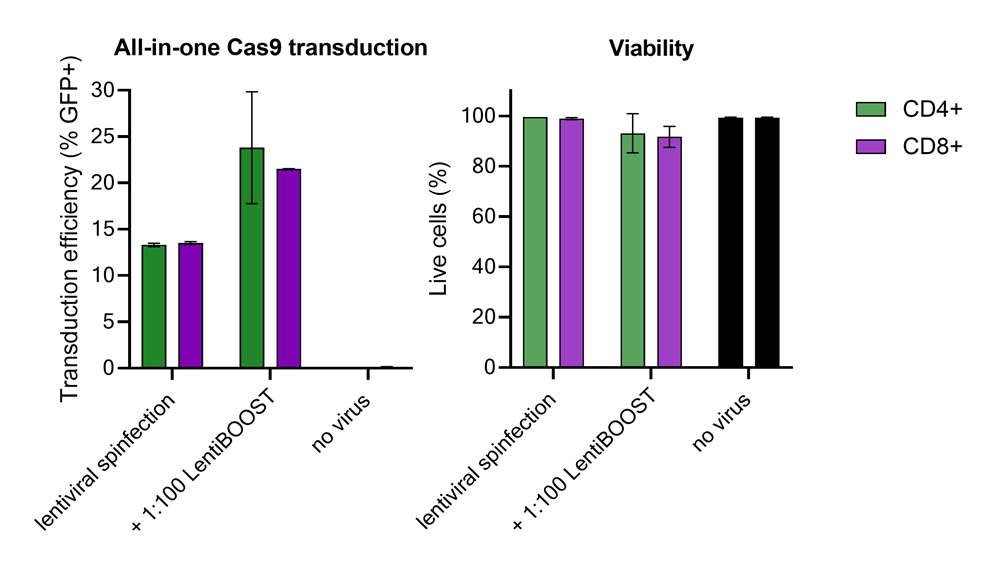
100,000 primary human CD4+ and CD8+ T cells from two donors were transduced with 250,000 TUs of Edit-R GFP Delivery controls mCMV along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved transduction of human induced pluripotent stem cells (hiPSCs) with the Strict-R™ Inducible CRISPRa lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer

10,000 WTC hiPS cells were transduced with either 20,000 (MOI 2, green) or 40,000 (MOI 4, purple) TUs of Strict-R™ Inducible EGFP dCas9-VPR Lentiviral Particles along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST markedly improved transduction efficiencies without significantly impacting cell viability.
Application notes
Posters
Protocols
Safety data sheets
Selection guides
Technical manuals
-
CRISPR-Cas9 genome engineering with inducible lentiviral Cas9 nuclease and Edit-R guide RNAs - Technical Manual
-
CRISPR-Cas9 genome engineering with lentiviral Cas9 particles and Edit-R synthetic guide RNA - Technical Manual
-
Edit-R CRISPR-Cas9 Gene Engineering with Lentiviral Cas9 and sgRNA - Technical Manual
Related Products
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.
Single guide RNA expressing vectors for effective and accurate gene knockout
Perform unbiased, phenotypic knockout screens without the need for costly infrastructure
Unlock precise, tunable gene knockout with the DharmaconTM Strict-RTM inducible Cas9 lentiviral system. This advanced platform enables stringent, control of gene disruption with exceptional specificity and minimal off-target activity. By integrating Tet-On 3G transcriptional regulation with FKBP12-derived degron–based destabilization, the system allows inducible expression and degradation of Cas9 in response to small-molecule regulators, providing tight temporal control over genome editing activity for dynamic and time-resolved functional studies.