- Gene modulation
- RNA interference
- siRNA solutions by Dharmacon™
- Lincode siRNA Reagents
Lincode siRNA Reagents
Knockdown of long noncoding RNA (lncRNA)
Expert siRNA design and specificity-enhancing modifications for effective knockdown of human long non-coding RNA. Predesigned Lincode siRNA is available as individual reagents and SMARTpool format.

Lincode siRNA Reagents
1Start Here
2Choose
Lincode siRNA reagents are an important tool for the study of long noncoding RNA (lncRNA); an emerging class of regulatory molecules whose roles are still being fully elucidated. The Lincode pre-designed human siRNA reagents benefit from expert siRNA design and proprietary dual-strand modifications for specificity. Simply search for your gene of interest and choose from the available product formats and quantities.
Highlights
- Pre-designed against 5867 characterized lncRNAs (NR_ and XR_ transcripts) in Human RefSeq v.65
- Lincode siRNAs carry proprietary dual-strand chemical modifications to ensure optimal strand loading for the reduction of off-targets
- SMARTpool reagent technology that targets multiple regions of the lncRNA to improve the likelihood of RNAi-mediated degradation
The importance of high-confidence siRNA reagents in lncRNA knockdown studies
lncRNA may be more difficult to silence than protein-coding genes due to:
- Nuclear localization of the lncRNA, reducing availability to RNAi cellular machinery
- Tertiary structure of the lncRNA, preventing siRNA access to the target region
- DNA or protein binding to the lncRNA, preventing siRNA access to the target region
Product Formats
- SMARTpool: A mixture of 4 siRNA provided as a single reagent; providing advantages in both potency and specificity.
- Set of 4: Discounted price on the Set of 4 siRNAs targeting a single gene. NO PRIOR PURCHASE REQUIRED.
- Individual siRNAs: For experiments where single-sequence application is required.
Endogenous lncRNA positive control and non-targeting control siRNA and pool reagents for high-confidence experiments. Lincode Control siRNA reagents incorporate specificity-enhancing modifications to improve experimental results.
Non-targeting siRNAs are designed with three or more mismatches to every lncRNA and protein-coding transcript in human, mouse, and rat. GAS5 is a widely expressed lncRNA whose knockdown has been demonstrated in multiple cell lines. If GAS5 expression levels are undetermined in your cell type, the use of ON-TARGETplus positive controls is an alternative method for initial experimental optimization.
| Lincode Positive Control siRNA | Species | Catalog Number |
|---|---|---|
| Lincode GAS5 Control siRNA | Human | D-001310-01 |
| Lincode GAS5 Control Pool | Human | D-001310-10 |
| Lincode Negative Control siRNA | Species | Catalog Number |
| Lincode Non-targeting Control siRNAs | Human, Mouse, Rat | D-001320-0X |
| Lincode Non-targeting Pool | Human, Mouse, Rat | D-001320-10 |
Lincode siRNAs effectively knockdown target lncRNAs
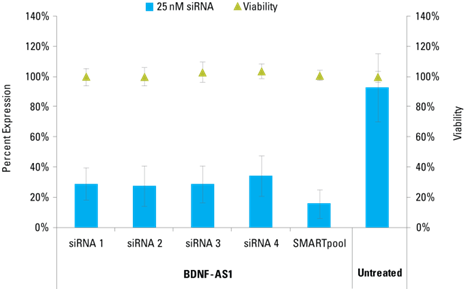
Knockdown of BDNF-AS1 in HeLa cells by Lincode SMARTpool and four siRNAs. All siRNAs were used at 25 nM, normalized and remaining lncRNA transcripts were detected with Solaris (sold under the Thermo Scientific brand) qPCR lncRNA Expression Assays and normalized to Non-targeting Control siRNA. Viability was assessed by resazurin assay.
Detection of effective lncRNA knockdown following application of Lincode siRNA
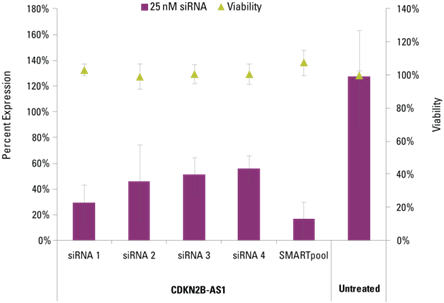
Knockdown of CDKN2B-AS1 in hNDF cells by Lincode SMARTpool and four siRNAs. All siRNAs were used at 25 nM, normalized and remaining lncRNA transcripts were detected with Solaris qPCR lncRNA Expression Assays and normalized to Non-targeting Control siRNA. Viability was assessed by resazurin assay.
lncRNAs demonstrate highly variable expression
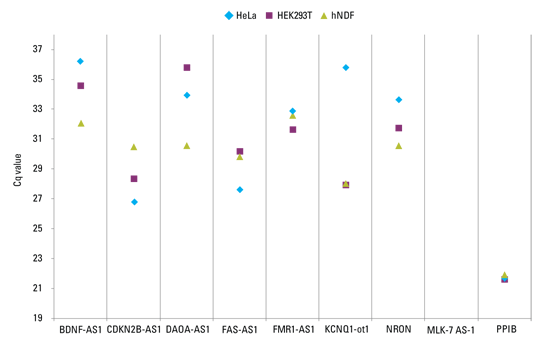
Robust qPCR assays are necessary for accurate determination of lncRNA expression levels.
lncRNAs demonstrate lower and more variable expression than protein-coding genes. It is therefore highly recommend to determine lncRNA expression levels in your cells prior to knockdown experiments. Solaris qPCR expression assays are designed for sensitive detection of lncRNA and demonstrate high sensitivity and reproducibility.
50 ng of total RNA was converted into cDNA using the Maxima First Strand cDNA Synthesis Kit (#K1641). Gene expression levels were evaluated with Solaris qPCR assays. Cyclophilin B (PPIB) was used as a reference for typical mRNA expression. Standard RT-PCR cycling conditions resulted in initial Cq values ranging from 22 (PPIB in 293T cells) to 36 (BDNF-AS1 in HeLa cells). MLK7-AS1 lncRNA expression was not detectable in any of the cell lines tested. Cell lines include HeLa, HEK293T, Human Neonatal Dermal Fibroblast (hNDF).
Lincode modifications enhance function and reduce off-targets
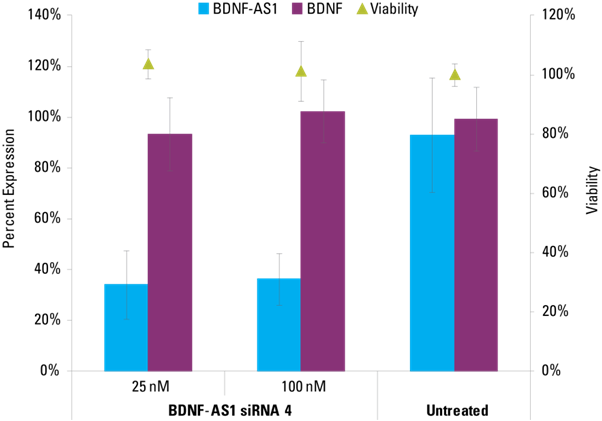
Lincode siRNA effectively knocks down BDNF-AS1 lncRNA, but not the protein-coding transcript BDNF that is antisense to the lncRNA target. This indicates strand specificity of Lincode siRNAs and effectiveness of ON TARGETplus modifications. siRNAs were applied at the indicated concentrations to hNDF cells. BDNF-AS1 and BDNF transcripts were detected with Solaris qPCR Expression Assays and normalized to Non-targeting control siRNA. Viability was assessed by resazurin assay and normalized to Untreated.
Lincode siRNA modifications greatly reduce Off-targets
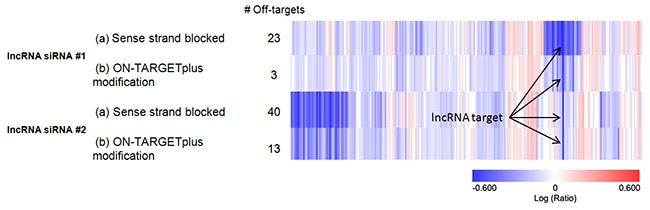
Genomewide expression analysis demonstrates improved specificity of Lincode siRNA
To detect subtle phenotypic changes that may arise from lncRNA knockdown, it is essential to incorporate strategies to prevent the off-targeting of protein-coding genes. Lincode siRNA reagents are synthesized with proprietary dual-strand modifications known as the ON-TARGETplus modification pattern which has been proven to reduce off-targets arising from microRNA-like activity of the antisense seed region of the siRNA.
Two different siRNAs targeting the same lncRNA were synthesized with modifications that (a) block the sense strand only or (b) the ON-TARGETplus modification pattern which blocks the sense strand and includes an antisense strand seed region modification. While the target lncRNA was effectively silenced by all four siRNAs (arrows), the siRNAs synthesized with the ON-TARGETplus modifications demonstrated greatly reduced off-targets.
Agilent™ G3 Human Gene Expression Microarray™ HeLa 12K, harvest 24 hrs post transfection, 100 nM siRNA, Analysis: >2 fold down regulated, pval <0.05, and non-siRNA-specific effects filtered out.
Sense strand activity is prevented by siRNA modifications
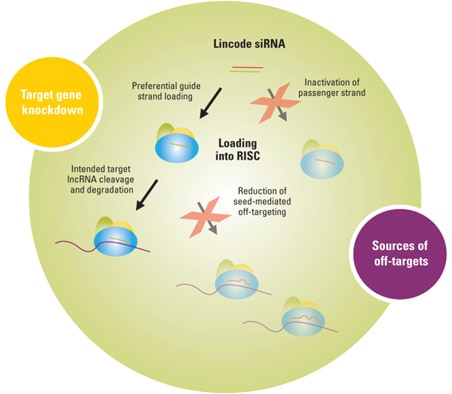
Lincode siRNAs are modified with a proprietary dual-strand modification that improves siRNA functionality. Additionally, off-targets are reduced due to:
- Inactivation of passenger strand activity; driving preferential loading of the guide strand into RISC
- Novel seed region modifications for disruption of microRNA-like off-targets
- C.P. Ponting, P.L. Oliver, Evolution and Functions of Long Noncoding RNAs. Cell 136, 629-641 (2009).
- L. Da Sacco, A. Baldassarre and A. Masotti. Bioinformatics Tools and Novel Challenges in Long Non-Coding RNAs (lncRNAs) Functional Analysis. Int. J. Mol. Sci. 13, 97-114 (2012)
Product inserts
Safety data sheets
Selection guides
Related Products
The most broadly applicable DharmaFECT formulation for optimal siRNA or microRNA transfection into a wide range of cell types for successful RNAi experiments
Catalog ID:T-2001-01
Unit Size:0.2 mL
$116.00
A positive control pool of four siRNAs targeting the GAS5 long noncoding RNA (lncRNA) in human cells. Includes patented dual-strand modifications to minimize off-target effects. Useful for determination of optimal RNAi conditions.
Catalog ID:D-001310-10-05
Unit Size:5 nmol
$269.00
Negative control pool of four siRNAs with three or more mismatches to any human, mouse or rat lncRNA or protein-coding gene. Dual-strand modifications reduce potential off-targets. Ideal for determination of baseline cellular responses in RNAi experiments.
Catalog ID:D-001320-10-05
Unit Size:5 nmol
