To stop or not to stop, but where is the question
The Precision LentiORF Collection
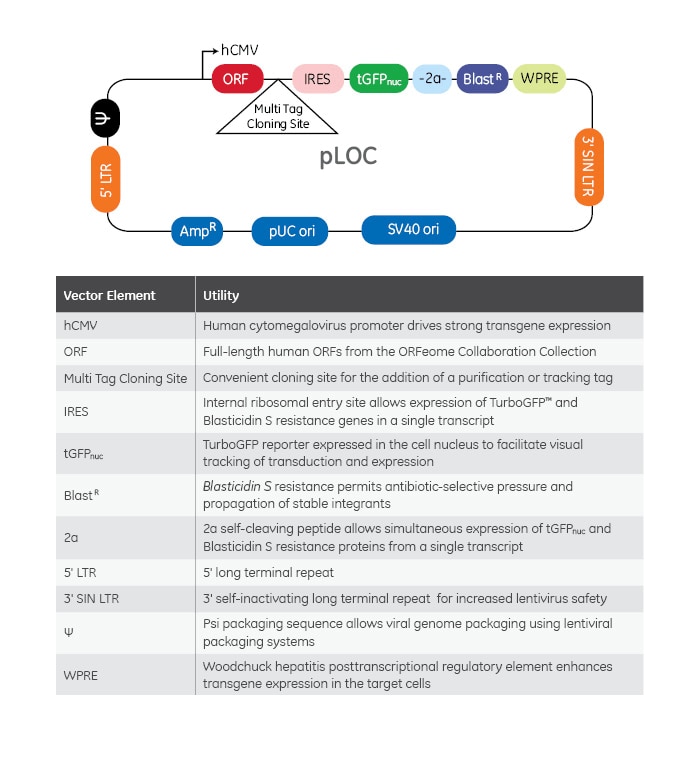
The Precision LentiORF Collection contains almost 12,000 human expression-ready lentiviral open reading frames (ORFs) generated from the original gene content of the ORFeome Collaboration Collection. The pLOC vector (Figure 1) has elements for expression of the fully sequenced ORF, a TurboGFP reporter, and a Blasticidin S resistance gene all from a single hCMV promoter. Utilization of a lentiviral vector allows packaging into lentiviral particles for delivery to difficult-to-transfect cells and creation of stable cells lines; while the single-promotor format helps maximize expression in your cell population and ensures that cells selected with either the resistance marker or fluorescence reporter are expressing the ORF sequence. There is also a Multi Tag Cloning Site (MTCS) immediately 3′ to the ORF coding sequence (more on this in a moment). Lastly, many Precision LentiORF clones are available either with or without the native stop codon. There is often confusion about what exactly is the utility of this great feature and when one should choose one format or the other for your experiments.
Figure 1. pLOC vector map
Ploc Vector Map Move Multitag NgPloc Vector Map Move Multitag Ng
Ploc Vector Map Move Multitag NgPloc Vector Map Move Multitag Ng
On the importance of a native stop codon
In order for a gene to be properly translated from an mRNA into a protein, it requires a stop codon; this is the signal that tells the ribosomal machinery to stop translation and end terminate the protein and release it into the cell to perform its function. If a gene sequence were to lack a stop codon, translation would span all the way to the 3′ end of the mRNA sequence and contain extra amino acids that would likely alter function. If a stop codon is introduced early in the sequence due to a mutation, the resultant protein would be truncated; again, with a likely alteration in function. A caveat to this is the ability to move the stop codon to a specific location in order to purposely manipulate the protein. This is what has been done with Precision LentiORF clones.
Selecting the best ORF format
For those clones that are described as “with native stop codon”, the stop codon for the ORF is in the characterized position at the C-terminus of the coding region (immediately before the vector’s MTCS). This means the ORF will be expressed and the protein translation end at the natural stop and no additional amino acids will be added. Use of these clones precludes access to the MTCS. Some applications where this type of ORF is commonly utilized are gene overexpression studies, RNAi rescue experiments, and structural analysis.
Clones that are “without native stop codon” can make use of the MTCS. In these clones, the native stop codon has been removed during the cloning process so the ribosome will read through the MTCS to a stop codon inserted on the vector backbone immediately downstream. The result is the addition of a few amino acids (approximately 13 - 15 ), or one has the option of inserting an additional sequence into this site, such as a 6x His or other similar epitope; easily creating C-terminal fusions. (Figure 2). This type of tagging can be advantageous if one is trying to visualize protein localization, purify biochemically, or identify interacting proteins through co-IP.
Figure 2. Highlighted gray are NheI and AscI restriction sites of the MCS.
Example of 3’ end of ORF and flanking sequence

Example of 3’ end of ORF and flanking sequence
