- Gene editing
- Gene editing reagents
- Edit-R predesigned lentiviral sgRNA
Edit-R predesigned lentiviral sgRNA
Single guide RNA expressing vectors for effective and accurate gene knockout
- Guaranteed to edit the target gene of interest
- Transduction-ready RNAs eliminate cloning and in vitro transcription steps
- Algorithm-optimized to maximize the likelihood of functional protein knockout and reduce off-target editing
- Available as glycerol stocks and high-titer purified particles

Edit-R predesigned lentiviral sgRNA
1Start Here
2Choose
Functional and specific targeting for high-confidence gene knockout results
Edit-R Lentiviral sgRNAs express RNA for guiding Cas9 nuclease to create double-strand breaks in the target DNA. In the Edit-R lentiviral sgRNA vector backbone, the gene-specific guide RNA is expressed under the control of a human U6 promoter, while expression of the puromycin resistance marker (PuroR) is driven from the mouse CMV promoter and allows for rapid selection of cells with integrated sgRNA.
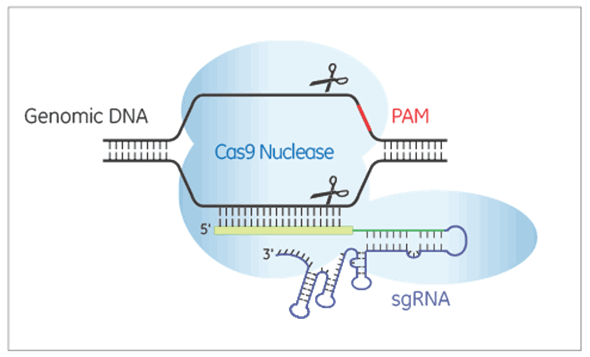
Each Edit-R Lentiviral sgRNA is specific to the gene and genomic site of interest. Generating knockouts in difficult-to-transfect cells or following up hits from a pooled lentiviral sgRNA library screen are key applications of this guide format.
While CRISPR-Cas9 is a highly effective tool for interrogating gene function, not all guide RNAs are effective in attaining functional protein knockout. To address this problem, Horizon developed an algorithm that is trained to select guides that give the highest likelihood of generating a functional gene knockout, not just creating an insertion or deletion.
New! Edit-R human sgRNA designs have been updated to the latest RefSeq in 2025 providing the most specific and genomically relevant guides for producing efficient protein knockout. This allows the Edit-R algorithm to target the latest genome annotations more accurately and efficiently providing you with the best solution for your research needs. Please reach out to Scientific Support if you have any questions or read our blog on revvity.com.
All guide RNA designs are top algorithm picks for each gene; qualitative ranks for functionality and specificity allow you to fine tune guide RNA choice to your specific application. The functionality score is a predicted indication of how likely this guide is to produce a functional knockout. The specificity score is based on the predicted risk of cutting activity at potential off-target sites. To learn more, visit our algorithm for Edit-R guide RNA page.
The Edit-R Predesigned Guide RNA Guarantee
We guarantee that EVERY predesigned guide RNA will provide successful editing at the target site when delivered as described in the Edit-R Technical Manuals.
The Edit-R guide RNA guarantee is valid when used with any wild type S. pyogenes Cas9 nuclease, including mRNA, expression plasmid, protein, or stable Cas9 expression, and Edit-R crRNAs must be used with Edit-R tracrRNA for the guarantee to apply.
Analysis of editing of the treated cell population must be shown using a T7EI or Surveyor mismatch detection assay. If successful editing is not observed for a predesigned Edit-R guide RNA while an appropriate side-by-side Edit-R positive control is successful, a one-time replacement of a different predesigned Edit-R guide RNA of the same format and quantity will be provided at no cost.
A replacement will only be approved upon discussion with our Scientific Support team.
Successful editing at the DNA level does not always lead to functional gene knockout; it is recommended to test multiple guide RNAs to determine the most effective guide RNA for knockout of your target gene.
This guarantee does not extend to any accompanying experimental costs, does not apply to guide RNAs ordered via the CRISPR Design Tool, and will not be extended to the replacement guide RNA.
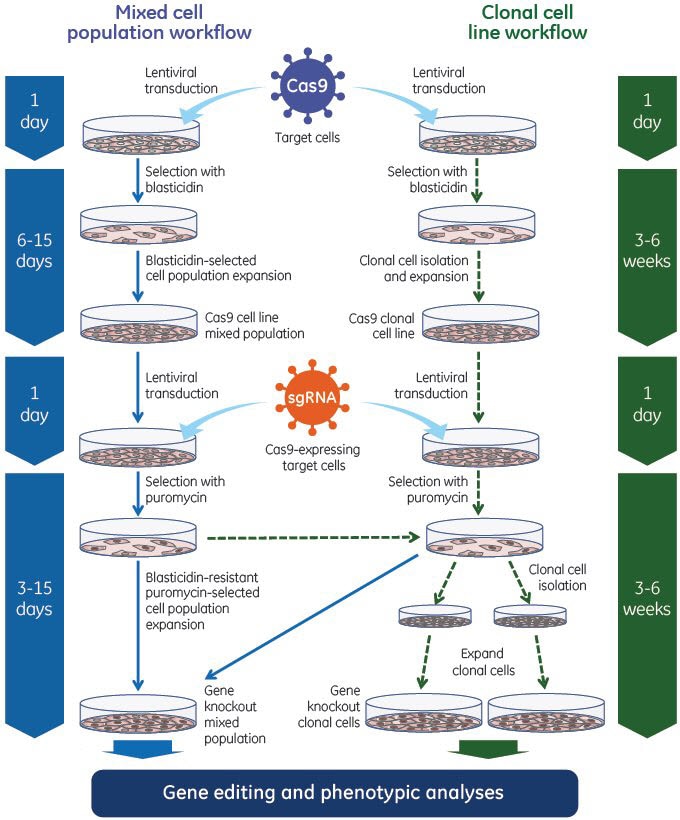
Experimental workflow using Edit-R Lentiviral sgRNA
The Edit-R Lentiviral Gene Engineering system utilizes Cas9 nuclease and the single guide RNA in a two-step process. First, Edit-R Lentiviral Cas9 Nuclease Expression particles are utilized to generate cell lines stably expressing Cas9 nuclease. These cells can subsequently be transduced with Edit-R Lentiviral sgRNA particles to achieve efficient gene editing - even at low MOIs - for phenotypic analyses in a population of cells or in isolated clonal cell lines.
Edit-R lentiviral sgRNA controls
-
Positive controls and detection primers
Species-specific sgRNAs targeting well-characterized genes to determine the effectiveness of your gene editing conditions for maximal efficiency.
-
Non-targeting controls
Lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human or mouse genomes.
-
Cutting controls
Lentiviral sgRNA constructs targeting inactive “safe harbor” genomic regions in the human and mouse genomes.
Edit-R lentiviral Cas9 under control of different promoters show high levels of gene editing in multiple cell lines with Edit-R Lentiviral sgRNA
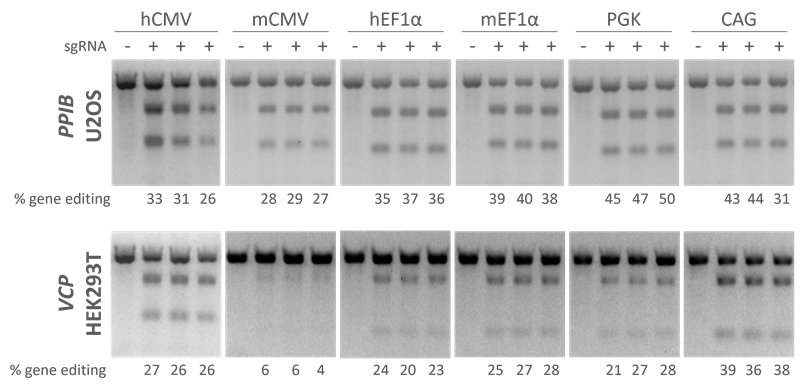
Testing gene editing with Cas9 under control of different promoters show that high levels of gene editing are achieved with Edit-R lentiviral sgRNA in multiple cell lines. A recombinant U2OS ubiquitin-EGFP proteasome cell line (Ubi[G76V]-EGFP) and HEK293T cells were stably transduced with lentiviral particles containing Cas9 and a blasticidin resistance gene. A population of stably integrated cells were selected with blasticidin for a minimum of 10 days before transduction with sgRNAs. Cells were transduced with sgRNA lentiviral particles at low MOI to obtain cells with one integrant and selected with puromycin for seven days prior to analysis. The relative frequency of gene editing in the puromycin-selected cells was calculated from a DNA mismatch detection assay using T7 Endonuclease I.
Schematic map of the plasmid vector elements of the Edit-R lentiviral sgRNA vector
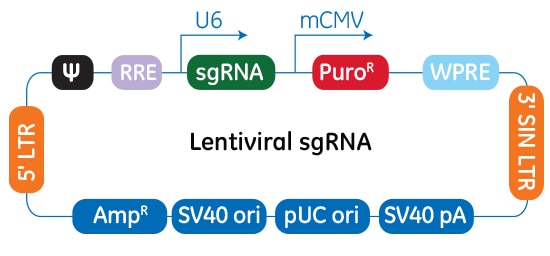
In the Edit-R lentiviral sgRNA vector backbone, the gene-specific crRNA and the tracrRNA are expressed under the control of a human U6 promoter, while expression of the puromycin resistance marker (PuroR) is driven from the mouse CMV promoter and allows for rapid selection of cells with integrated sgRNA. The plasmid contains the AmpR resistance marker for growth and selection in E. coli.
Algorithm applies to both synthetic crRNAs and expressed sgRNAs
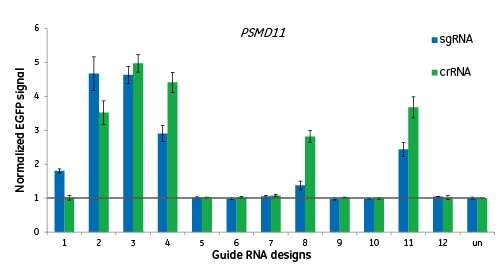
Here we are targeting the gene PSMD11 at 12 different sites and measuring functionality by EGFP signal to indicate disruption of the proteasome. Cells were either transfected with crRNA:tracrRNAs, or transduced with lentiviral particles for expression of a sgRNA of the same design. There is no significant difference between the two guide RNA formats when targeting the same genomic site; a good design for crRNA will translate into a good design for sgRNA.
Successful CRISPR-Cas9 gene editing utilizing co-delivery of Edit-R lentiviral sgRNA and Cas9 expression plasmid using DharmaFECT kb transfection reagent
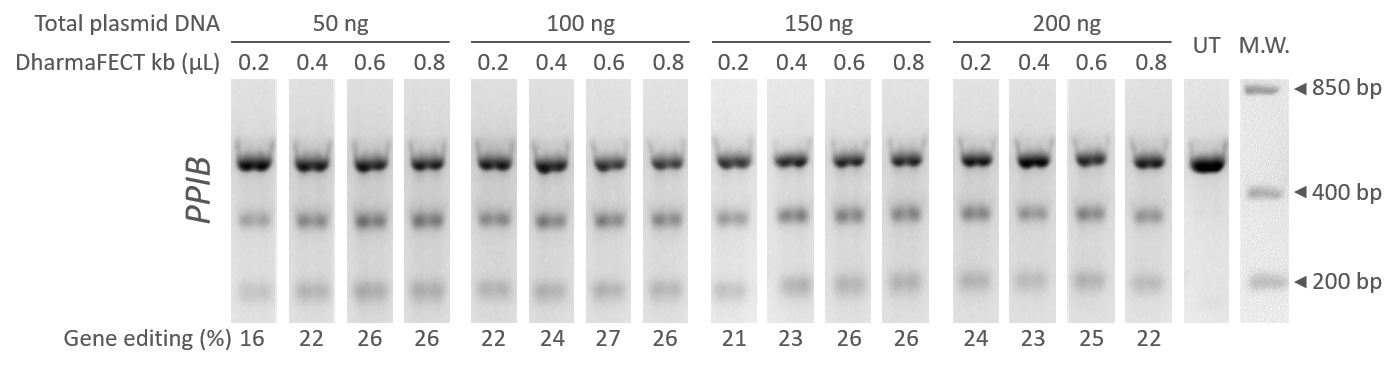
U2OS cells were transfected with equal amounts of Edit-R hCMV-mKate2-Cas9 Expression plasmid (Cat #U-004100-120) and Edit-R Human PPIB lentiviral sgRNA plasmid in a 96-well format. Transient transfections were done with increasing amounts of total DNA (50 to 200 ng) and DharmaFECT kb transfection reagent (Cat #T-2006-01), 0.2 to 0.8 µL per well. The percentage of gene editing was estimated 72 hours after transfection by DNA mismatch detection assay using T7EI with gel densitometry.
LentiBOOST Lentivirus Transduction Enhancer is a uniquely formulated transduction reagent that can be used with or without lentivirus spinfection in order to increase successful viral transduction events while preserving cell viability. Especially critical for preserving precious primary cells from patient cohorts, or, for engineering complex animal models; improving transduction efficiency can save time and costs by increasing the success of each editing/transduction step, or, even avoid the loss of irreplaceable samples. Additionally, LentiBOOST technology is already used in the manufacturing of a number of clinical stage therapies providing the opportunity to demonstrate improved workflow applicability to the clinic.
LentiBOOST can be purchased through the Dharmacon Reagents catalog.
To learn more about LentiBOOST technology visit the Revvity LentiBOOST webpage.
Supporting Data
Improved CD8+ T-cell SMARTvector™ shRNA lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
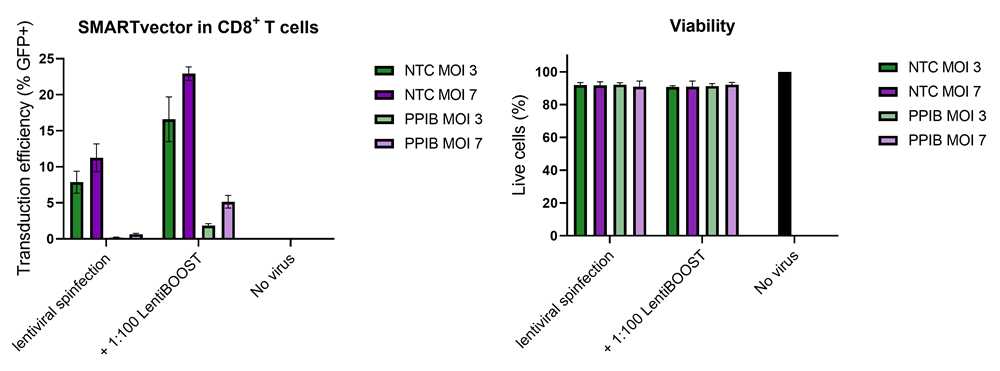
100,000 primary human CD8+ T cells were transduced with either 30,000 (MOI 3, green) or 70,000 (MOI 7, purple) TUs of SMARTVector™ mCMV tGFP Lentiviral Control Particles targeting either NTC or PPIB along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by a four hour incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency (%GFP+ out of live cells) and viability were determined 5 days post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved CD4+ and CD8+ T-cell Edit-R™ All-in-one sgRNA/Cas9 lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
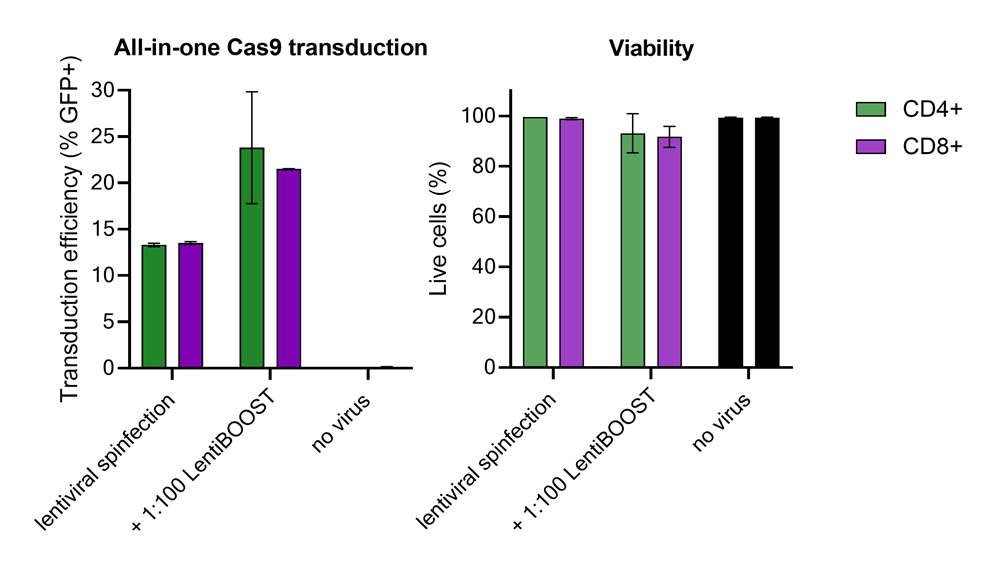
100,000 primary human CD4+ and CD8+ T cells from two donors were transduced with 250,000 TUs of Edit-R GFP Delivery controls mCMV along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved transduction of human induced pluripotent stem cells (hiPSCs) with the Strict-R™ Inducible CRISPRa lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer

10,000 WTC hiPS cells were transduced with either 20,000 (MOI 2, green) or 40,000 (MOI 4, purple) TUs of Strict-R™ Inducible EGFP dCas9-VPR Lentiviral Particles along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST markedly improved transduction efficiencies without significantly impacting cell viability.
Additional Resources
Application notes
Posters
Protocols
Safety data sheets
Selection guides
Related Products
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.
Lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human or mouse genomes.
Control lentiviral sgRNAs to verify DNA double-strand breaks and gene editing efficiencies.
Perform unbiased, phenotypic knockout screens without the need for costly infrastructure
