- Gene editing
- Gene editing reagents
- Edit-R Lentiviral sgRNA Positive Controls and Kits
Edit-R lentiviral sgRNA positive controls and kits
Control lentiviral sgRNAs to verify DNA double-strand breaks and gene editing efficiencies.
Validated Edit-R Lentiviral sgRNA Positive Controls and kits confirm generation of insertions and deletions (indels), and permit quantification of gene editing efficiency using DNA mismatch detection assays.

Validated Lentiviral sgRNA Positive Controls and Kits for the verification of gene editing events
Edit-R Lentiviral sgRNA Positive Controls and kits are designed to target human or mouse genes and can be included in all gene editing experiments to confirm successful transduction by observing the generation of insertions and deletions (indels) in your targeted genomic DNA using simple DNA mismatch detection assays.
Individual Edit-R Lentiviral sgRNA Positive Controls are available as glycerol stocks (mouse designs only) and as concentrated, purified lentiviral particles (50 µL at ≥ 1 x 108 TU/mL); available for human or mouse and designed against:- Cyclophilin B (PPIB)
- DNA (cytosine-5)-methyltransferase 3B (DNMT3B)
Edit-R Lentiviral sgRNA Positive Control Kits include:
- Lentiviral particles or glycerol stock (mouse designs only)
- DNA mismatch detection primers (forward and reverse)
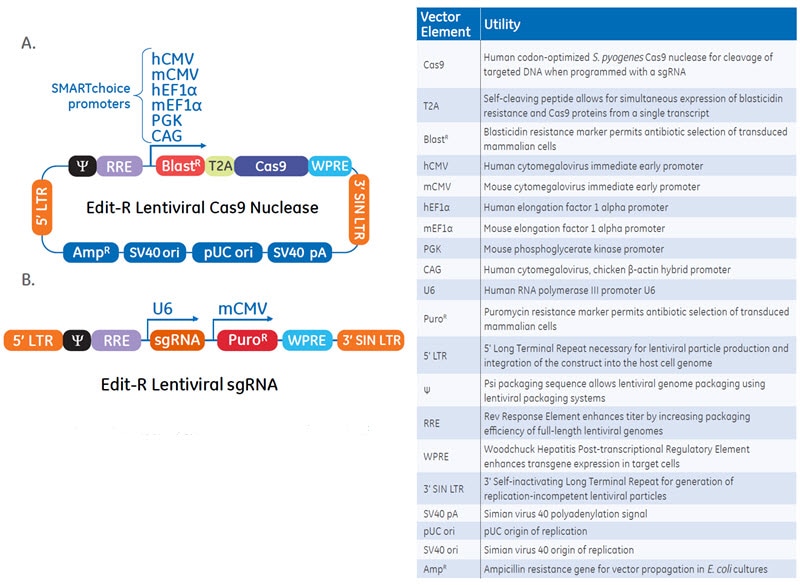
The Edit-R Lentiviral CRISPR-Cas9 Gene Engineering platform includes two critical components based on the natural S. pyogenes system: a vector for Cas9 expression and a gene-specific vector for sgRNA expression designed to the target site of interest. To facilitate rapid generation of cells lines that constitutively express Cas9 nuclease and the sgRNA, the Cas9 and sgRNA vectors are available pre-packaged into lentiviral particles that have been purified and concentrated. Cas9-expressing cells lines can be easily generated with the Edit-R Lentiviral Cas9 Nuclease Expression particles and subsequent gene knockouts can be obtained by an additional transduction with sgRNA lentiviral particles.
Schematic maps and table of vector elements of the Edit-R Lentiviral Cas9 Nuclease (A) and sgRNA (B) vectors
The Edit-R Lentiviral CRISPR-Cas9 platform is a two-vector system that utilizes a lentiviral vector with multiple Pol II promoter options for maximal Cas9 expression and a gene-specific vector for sgRNA expression designed to the target site of interest. In the Edit-R Lentiviral sgRNA vector backbone, the gene-specific crRNA and the tracrRNA are expressed under the control of a human U6 promoter, while expression of the puromycin resistance marker (PuroR) is driven from the mouse CMV promoter and allows for rapid selection of cells with integrated sgRNA.
Schematic map of the plasmid vector elements of the Edit-R Lentiviral sgRNA vector
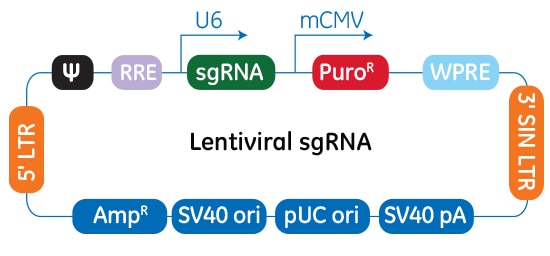
In the Edit-R Lentiviral sgRNA vector backbone, the gene-specific crRNA and the tracrRNA are expressed under the control of a human U6 promoter, while expression of the puromycin resistance marker (PuroR) is driven from the mouse CMV promoter and allows for rapid selection of cells with integrated sgRNA. The plasmid contains the AmpR resistance marker for growth and selection in E. coli.
Using Edit-R Lentiviral sgRNA Positive controls to detect varying levels of gene editing in HEK293T cells due to differential expression of Cas9 under the control of different promoters
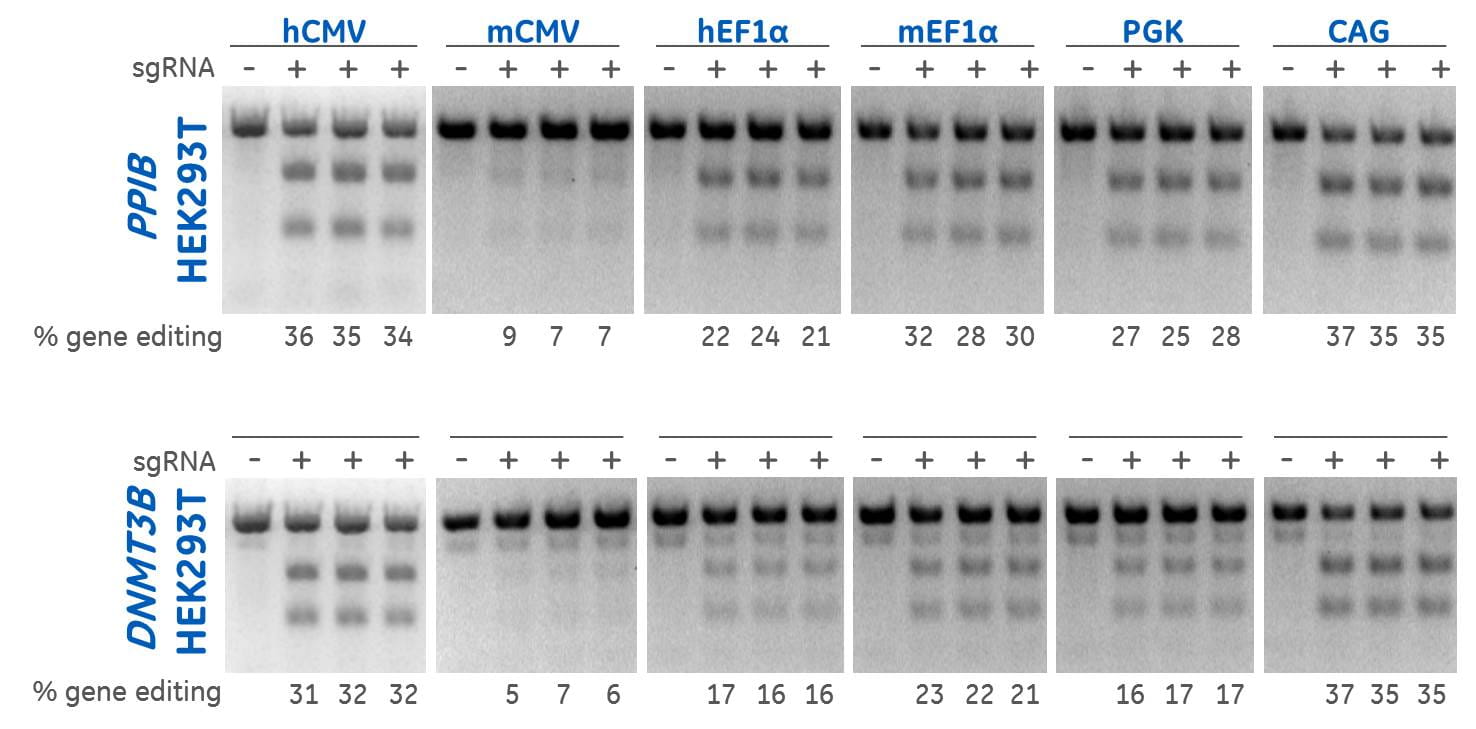
Using Edit-R Lentiviral sgRNA Positive controls and a mismatch detection assay to test efficiency of gene editing in HEK293T cells show that the hCMV and CAG promoters result in the highest levels of editing. HEK293T cells were stably transduced with lentiviral particles containing Cas9 and a blasticidin resistance gene. A population of stably integrated cells were selected with blasticidin for a minimum of 10 days before transduction with sgRNAs. Cells were transduced with positive control sgRNA lentiviral particles at low MOI to obtain cells with one integrant and selected with puromycin for 7 days prior to analysis. The relative frequency of gene editing in the puromycin selected cells was calculated from DNA mismatch detection assay using T7 Endonuclease I.
Application notes
Protocols
Safety data sheets
Related Products
Single guide RNA expressing vectors for effective and accurate gene knockout
Lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human or mouse genomes.
Purified lentiviral particles for generation of stable Cas9 nuclease-expressing cell populations
Perform unbiased, phenotypic knockout screens without the need for costly infrastructure
