- Gene editing
- Gene editing reagents
- Edit-R lentiviral sgRNA negative controls
Edit-R lentiviral sgRNA negative controls
Lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human or mouse genomes.
Negative control sgRNAs to establish experimental baselines and to distinguish sequence-specific biological effects from non-specific effects. Available as high-titer purified lentiviral particles and glycerol stock formats (mouse designs only).

AAVS1/Rosa26 cutting controls are designed to target regions of the genome that do not result in functional knockout of a protein or known phenotypic changes. These regions are sometimes known as safe harbor regions. These guides are validated for mismatch detection assays and are recommended for screening applications as an additional control that results in Cas9 cutting without a phenotypic readout.
Non-targeting controls are recommended as negative controls for experiments using synthetic sgRNA. All Edit-R non-targeting controls are designed to have a minimum of three mismatches or gaps to all potential PAM-adjacent targets in the human or mouse genomes. Changes in viability or gene expression levels in cells treated with these controls likely reflect a baseline cellular response that can be compared to the levels in cells treated with target-specific sgRNAs.
Edit-R CRISPR-Cas9 Gene Engineering Platform
The Dharmacon Edit-R Gene Engineering platform is powered by a proprietary algorithm and based on the Type II CRISPR-Cas9 system from the bacteria Streptococcus pyogenes.
- Ready-to-Use Components: The system eliminates the need for cloning, allowing researchers to quickly initiate experiments with predesigned guide RNAs and other essential components, significantly reducing the time to results.
- High Specificity and Efficiency: Edit-R utilizes a proprietary algorithm to design guide RNAs that improve the likelihood of achieving functional knockouts while minimizing off-target effects, ensuring high confidence in experimental outcomes.
- Guaranteed Performance: Every predesigned guide RNA is backed by a guarantee of successful editing at the target site, providing researchers with additional assurance in their gene editing endeavors.
The Edit-R predesigned guide RNA guarantee
We guarantee that EVERY predesigned guide RNA will provide successful editing at the target site when delivered as described in the Edit-R Technical Manuals.
The Edit-R guide RNA guarantee is valid when used with any wild type S. pyogenes Cas9 nuclease, including mRNA, expression plasmid, protein, or stable Cas9 expression, and Edit-R crRNAs must be used with Edit-R tracrRNA for the guarantee to apply.
Analysis of editing of the treated cell population must be shown using a T7EI or Surveyor mismatch detection assay. If successful editing is not observed for a predesigned Edit-R guide RNA while an appropriate side-by-side Edit-R positive control is successful, a one-time replacement of a different predesigned Edit-R guide RNA of the same format and quantity will be provided at no cost.
A replacement will only be approved upon discussion with our Scientific Support team.
Successful editing at the DNA level does not always lead to functional gene knockout; it is recommended to test multiple guide RNAs to determine the most effective guide RNA for knockout of your target gene.
This guarantee does not extend to any accompanying experimental costs, does not apply to guide RNAs ordered via the CRISPR Design Tool, and will not be extended to the replacement guide RNA.
Gene knockout workflow using the Edit-R All-in-one lentiviral sgRNA system
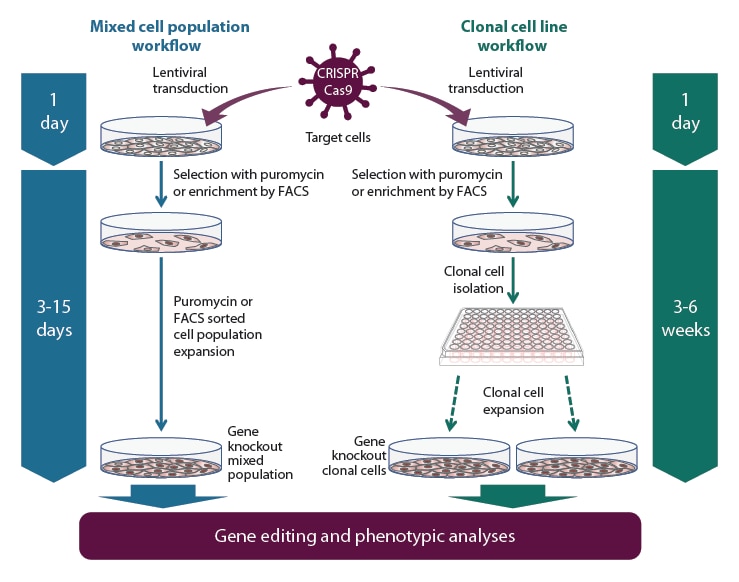
Gene knockout workflow with the All-in-one lentiviral sgRNA using a mixed popultion (left side) or clonal cell line (right side) experimental approach.
Schematic maps and table of vector elements of the Edit-R Lentiviral Cas9 Nuclease and sgRNA vectors
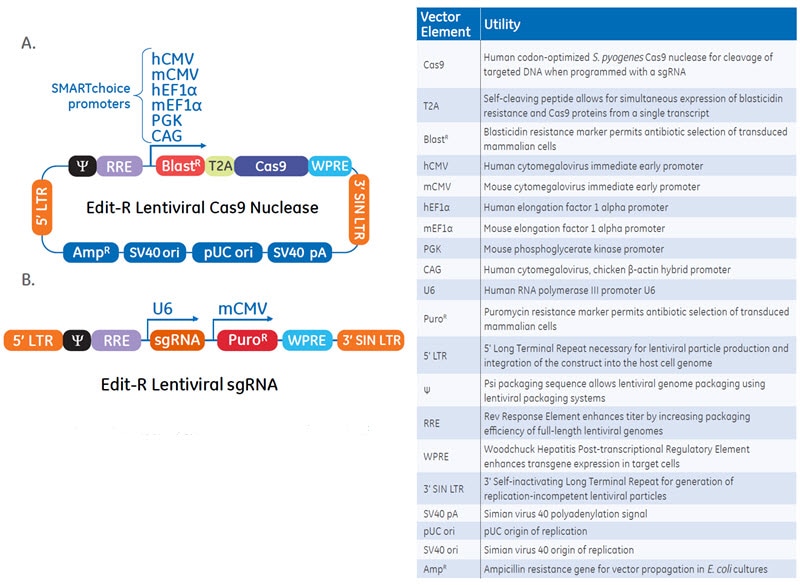
Schematic maps and table of vector elements of the Edit-R Lentiviral Cas9 Nuclease and sgRNA vectors.The Edit-R Lentiviral CRISPR-Cas9 platform is a two-vector system that utilizes a lentiviral vector with multiple Pol II promoter options for maximal Cas9 expression and a gene-specific vector for sgRNA expression designed to the target site of interest.
Schematic map of the plasmid vector elements of the Edit-R Lentiviral sgRNA vector
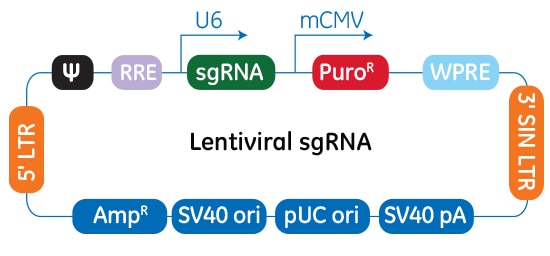
In the Edit-R Lentiviral sgRNA vector backbone, the gene-specific crRNA and the tracrRNA are expressed under the control of a human U6 promoter, while expression of the puromycin resistance marker (PuroR) is driven from the mouse CMV promoter and allows for rapid selection of cells with integrated sgRNA. The plasmid contains the AmpR resistance marker for growth and selection in E. coli.
Experimental workflow using Edit-R Lentiviral sgRNA
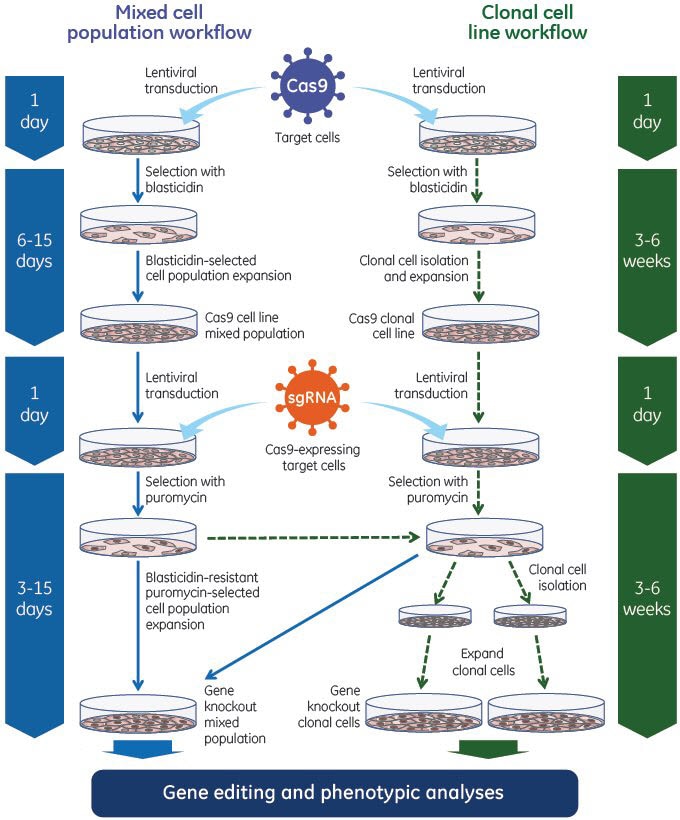
Experimental workflow using Edit-R Lentiviral sgRNA. Edit-R Lentiviral sgRNA are transduced into a stable cell line expressing Cas9 nuclease for efficient gene knockout, even at low MOIs.
Protocols
Safety data sheets
Related Products
Single guide RNA expressing vectors for effective and accurate gene knockout
Control lentiviral sgRNAs to verify DNA double-strand breaks and gene editing efficiencies.
Purified lentiviral particles for generation of stable Cas9 nuclease-expressing cell populations
Perform unbiased, phenotypic knockout screens without the need for costly infrastructure
