CRISPR technology now allows genes and molecular pathways to be examined with greater definition. We look at how knockout cell lines, either together with gene rescue and replication of disease mutations or as an independent cell model, can be used to validate your research and extend your findings.
There is a significant challenge in translating the wealth of genetic information now available to the role of genes, to understand basic biology, as well as linking to the role of mutations for understanding disease pathogenesis.
Knockout (KO) cell lines are excellent model systems to do this. A key benefit of cell lines is the ability to use gene-editing to construct isogenic cell line pairs - where a mutant model can be interrogated alongside a wildtype control.
Here's five great examples of how to get the most out of your research using KO Cell Lines:
- Validate your research tools
- Identify key players in a pathway
- Confirm pharmacological effects
- Lift the quality of your research with proper controls
- Connect patient relevant mutations to pathology
Antibody validation
We know that the quality of research tools used impacts significantly on the quality of research delivered. Antibodies are one of the most commonly used reagents in life-sciences, but despite their widespread use, there are no standard guidelines for how these invaluable biological tools should be validated prior to use.
For example, poorly characterized antibodies may yield non-specific results, which are difficult to replicate even when the only difference is the antibody's production lot. However, the bar for antibody validation is rising and there are calls for the industry standard to show application-specific validation using KO cell lines.
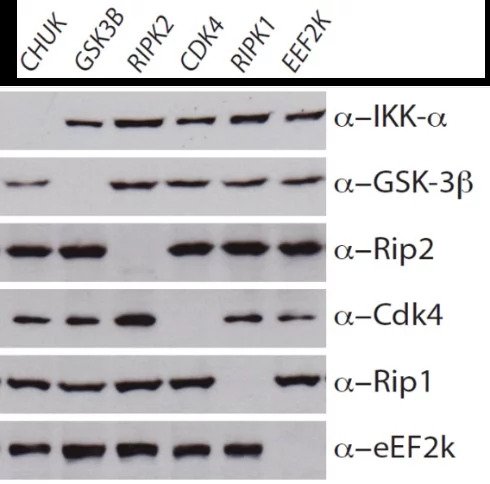
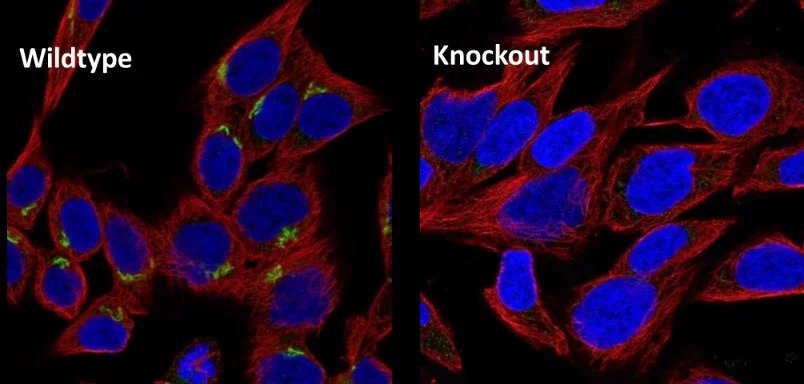
This example shows data from a selection of KO cell lines. The western blot data confirms absence of protein, and so validates the antibody used. Similarly, in the IHC image, the absence of green staining validates the specificity of the antibody used here. This shows that parallel use of wildtype and KO cell lines provides a valuable tool to control for research reagents quality.
Pathway interrogation with controls for off-target effects
Often there are multiple related, but distinct molecules and processes present in a pathway. Gene-edited cell lines are a well-established tool to probe the role of a particular gene or mutation.
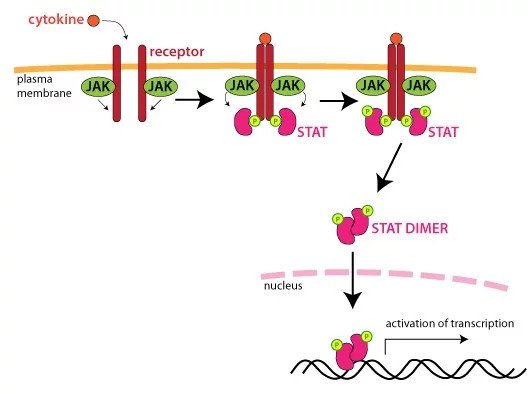
Here is an example of how HAP1 KO cell lines can be used to tease out the role of a protein in a pathway.
JAK1 is essential for signaling for certain cytokines and is thought to play a role in tumor metastasis. It is known to have a functional and physical association with INF gamma receptors. Upon ligand binding, receptor-associated JAKs become activated, leading to the recruitment of specific STATs (signal transducer and activator of transcription) from the cytoplasm.
These STATs then become JAK substrates. Activated STATs are released from the receptor, which then translocate to the nucleus to bind to specific enhancer elements. These then affect the expression of target genes.
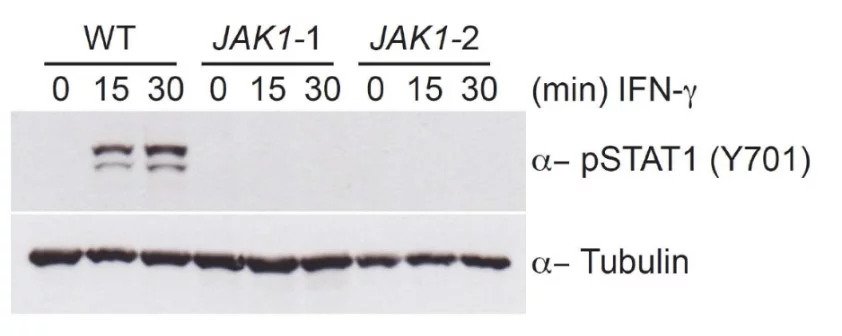
The role of JAK1 in STAT phosphorylation can be confirmed by creating a frameshift mutation in the JAK1 gene, in this case in theHAP1 cell line. These KO cell lines do not allow for the phosphorylation of STAT, as can be seen in this western blot.
To mitigate against any effects of off-target editing in CRISPR-Cas9 engineered cell lines, it is useful to use KO cell line clones created by two different sgRNAs. If off-target editing occurs, this will then show at different sites, so you can have confidence that any common phenotypes between the two lines are due to mutations in your target gene only. In this experiment, two separate HAP1 clones were used to confirm these findings, indicating this was not an off-target effect.
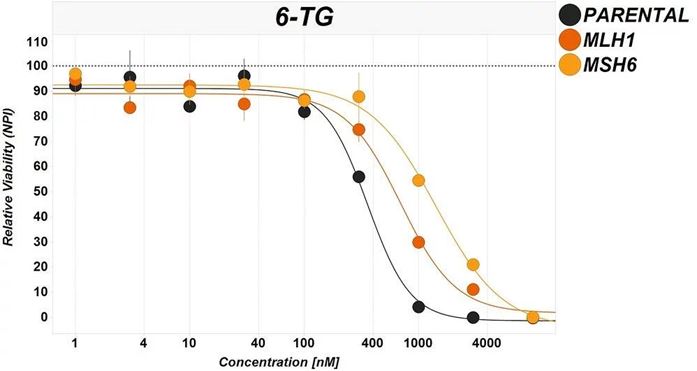
Pharmacological interventions are commonly used to interrogate pathways. In this example the chemotherapeutic 6-thioguanine (6-TG) is used to probe the effect of DNA damage response. To maintain genomic integrity, cells are equipped with a myriad of mechanisms that are each specific for different types of damage. The efficiency of these DDR pathways plays an essential role in the confirmation of pharmacological effect and effectiveness of cytotoxic treatments.
6-TG is a growth inhibitory antimetabolite which requires an active DNA mismatch repair (MMR) system to be effective. KO cell lines which are devoid of key proteins are used to confirm the target of the 6-TG.
Here, KO cell lines for MLH1 or MSH6 confer resistance to the cytotoxic effects of 6-TG, confirming the involvement of the MMR pathway. You can see details of the full experiment and results in this blog.
Rescue and disease models
After identifying mutations of interest in a patient population, the next steps are to probe the biological mechanisms that result in the disease phenotype or the response to treatment.
KO cell lines, combined with simple molecular biology techniques, allows you to:
- Rescue the gene of interest to restore phenotype
- Complement the Knockout cell line with a functional mutation, such as a point mutation observed in a patient population
This example involves the role of an E3 ligase called RFWD3. E3 ubiquitin ligases are known to have a role in DNA damage repair, particularly in homologous recombination (HR). BRCA1 encodes an E3 ligase that is critical for HR and defects in the BRCA1 protein often lead to breast cancer.
In this example from the Takata Group, the key to clarifying the role of RFWD3 in DNA damage repair was obtaining a system with complete loss of function of that particular E3 ligase, to isolate its specific role from potentially overlapping molecules. HAP1 KO cell lines are a simple tool that were used to accomplish this.
Here are survival curves of HAP1 WT, KO RFWD3, or KO RFWD3 stably transduced with a lentivirus expressing WT or one of 2 RFWD3 mutations identified in patient populations.
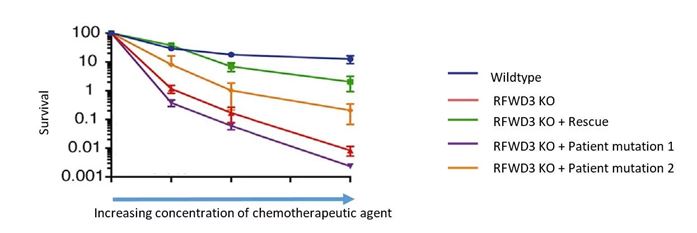
The KO RFWD3 cells exhibited hypersensitivity to MMC (mitomycin C), whereas reintroduction of wild-type RFWD3 into the KO model reversed MMC sensitivity. However, introduction of patient relevant point mutations in to the KO model showed distinct responses. One mutations demonstrate partial rescue, whereas a separate point mutation, (a RING finger domain mutant - C315A) did not.
Connecting patient relevant mutations to pathology
KO cell lines are a valuable resource to connect findings from a patient population to the role of a specific gene. This example links regulators of the maintenance of functional chromatin domains to docetaxel response. First, gene expression data, generated from breast cancer patient derived xenograft (PDX) models and patient samples treated with docetaxel, was used to identify the involvement of a set of chromatin regulators.
Results show that specific chromatin regulators, including key components of the SWI/SNF chromatin remodeler, readily distinguish docetaxel high-responders and poor-responders. By examining dataset of 60 diverse human cancer cell lines, the expression of SWI/SNF components was shown to inversely correlate with docetaxel sensitivity.
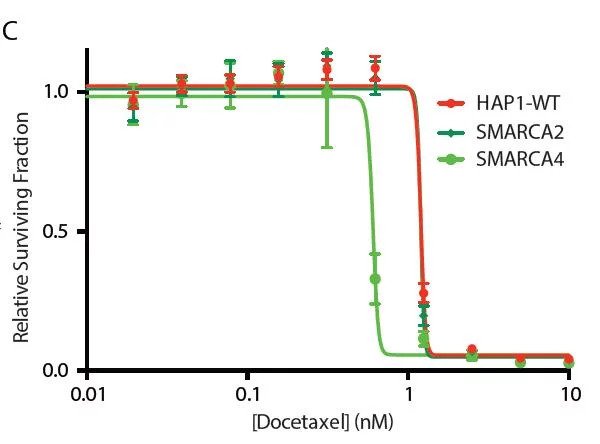 Confirmation of these findings was possible by using a HAP1 KO cell line, showing loss of the SWI/SNF subunit BRG1 (SMARCA4) in a model cell line leads to enhanced docetaxel sensitivity.
Confirmation of these findings was possible by using a HAP1 KO cell line, showing loss of the SWI/SNF subunit BRG1 (SMARCA4) in a model cell line leads to enhanced docetaxel sensitivity.
The results show a 4-fold decrease in the IC50 for docetaxel in the SMARCA4-KO cell line, relative to wildtype. These findings point towards chromatin regulators as biomarkers for drug response as well as therapeutic targets to sensitize patients towards docetaxel and combat drug resistance.
Summary
Hap1 KO Cell Lines are a versatile tool for helping us understand basic biology, as well as identifying the role of mutations in disease pathogenesis. Ultimately, this has led to advances in disease treatment and patient prognosis.
Horizon has a vast range of Knockout Cell Lines to elevate your research.
Order Horizon HAP1 KO cell lines
Learn more
- Is your antibody binding the right target? – Featured article
- HAP1 cell lines - are they the right cell model for you? – Blog article
- Frequently asked questions for HAP1 knockout cell lines – Blog article
