Based on critical components of the well-studied CRISPR-Cas9 system, the Edit-R™ All-in-one Lentiviral sgRNA vector rapidly generates stable cell lines for phenotypic analysis of gene knockout.

What is the Edit-R All-in-one CRISPR system?
The system is a Lentiviral sgRNA vector including all necessary components for S. pyogenes CRISPR-Cas9 gene editing. Each sgRNA vector contains a gene-specific single guide RNA (sgRNA) and Cas9 nuclease. Additionally, the vector is available in two configurations enabling antibiotic resistance or fluorescence sorting. It is supplied packaged into lentiviral particles for immediate transduction of mammalian cells grown in culture. Both pre-designed and custom targeting sequences are available for knockout of specific target genes, with expansion of the resultant stable cell lines for phenotypic analysis.
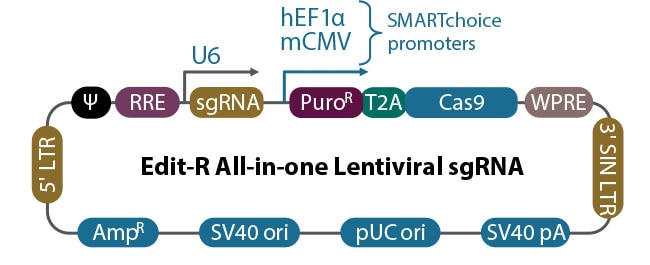
Figure 1. Schematic of the Edit-R All-in-one Lentiviral sgRNA vector.
An easy to use, single reagent for CRISPR-Cas9 gene knockout
A main advantage of using the Edit-R All-in-one Lentiviral sgRNA vector for CRISPR-Cas9 gene editing is the simple experimental workflow. After thawing the lentivirus and pipetting an optimized amount onto cells grown in culture, researchers simply perform selection with puromycin, or isolation and enrichment by fluorescence-activated cell sorting (FACS), to obtain stable gene knockout cells.
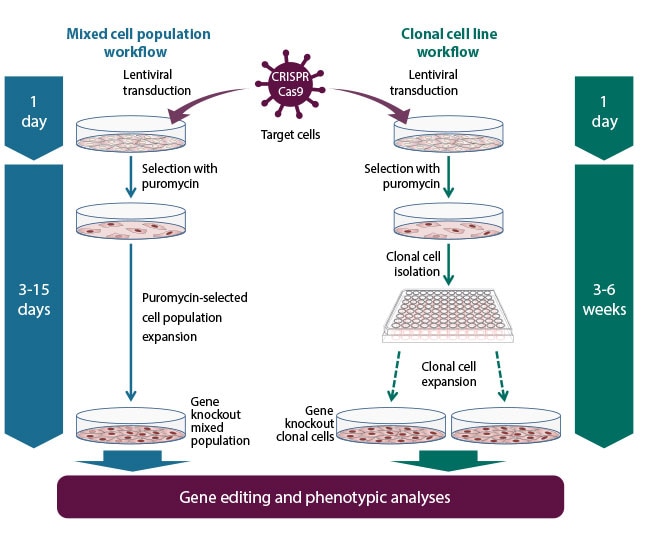
Figure 2. Gene knockout workflow using the Edit-R All-in-one Lentiviral sgRNA system. Gene editing with Edit-R All-in-one Lentiviral sgRNA can follow the mixed cell populations approach (left) typically for gene knockout screenings, or the isolated clonal cell lines approach (right) when a defined genotype is desired or required on each step for phenotypic analyses.
Lentiviral delivery enables gene knockout for "difficult-to-transfect" cells
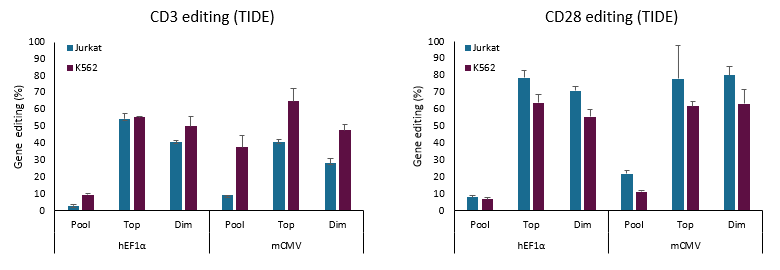
Another important benefit of the Edit-R All-in-one Lentiviral sgRNA system is that it enables gene knockout in mammalian cells renowned for being difficult-to-transfect. In a direct comparison between K562 and Jurkat (a difficult to transfect cell line which mimics T cells), similar editing percentages as shown by TIDE analysis were seen in both cell lines using Edit-R All-in-one EGFP Lentiviral sgRNA vectors to target CD3 or CD28 genes.
Figure 3. Gene knockout workflow using the Edit-R All-in-one Lentiviral sgRNA system. Gene editing with Edit-R All-in-one Lentiviral sgRNA can follow the mixed cell populations approach (left) typically for gene knockout screenings, or the isolated clonal cell lines approach (right) when a defined genotype is desired or required on each step for phenotypic analyses.
Save time using selectable markers to enrich for edited cells
In addition to sgRNA and Cas9, the Edit-R All-in-one Lentiviral sgRNA vector includes several other key elements. The puromycin resistance (PuroR) and EGFP markers save researchers’ time by permitting antibiotic selection or identification and enrichment of transduced cells by FACS, respectively.
Perform double knockouts with multiple selection markers
By using both PuroR and EGFP selection markers, the Edit-R All-in-one Lentiviral sgRNA vector enables double knockouts. Knocking out multiple genes helps unravel complex cellular signaling pathways and identify redundancy within the underlying biology.
 Written by Ryan Donnelly
Written by Ryan Donnelly
He holds a Professional Science Masters in Molecular Biotechnology from George Washington University and Bachelors in Chemistry from the University of Florida.
To learn more about the Edit-R All-in-one Lentiviral sgRNA vector, take a look at our applications page.
Ready to get your next experiment started?
If you have any further questions or need some help troubleshooting your experiment, our Scientific Support team is available to provide expert guidance and support.