HAP1 cell origins and how this model can help your research
Horizon HAP1 knockout cell lines are the solution to quickly assess phenotype without the time and screening needed to produce a single selected clone in your lab.
Great for quick proof of concept experiments, assistance with antibody validation, or directly answering the question if a knockout cell line of the gene of interest will produce the desired data.
This article will go through common questions our Scientific Support team receives on the HAP1 cells, including:
• What is the origin of the HAP1 cells?
• How are these knockouts produced?
• What quality control steps are taken in the production of these cell lines?
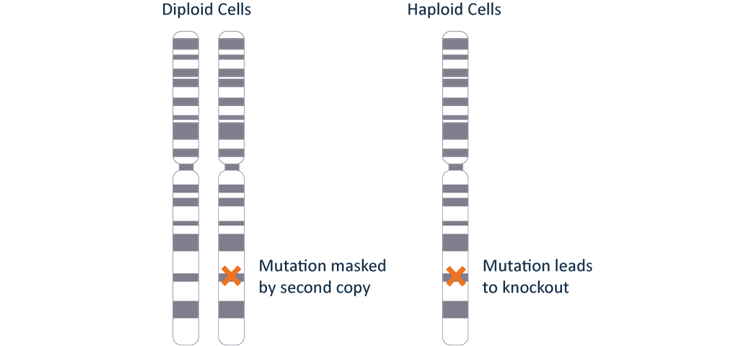
What is the benefit of a knockout produced in a haploid cell line?
Most normal human cells are diploid; they have two sets of chromosomes. Cancer cell lines and immortalized cultured cell lines often have additional copies of genes. When analyzing a cell line in which to perform a knockout one can quickly realized many cell lines in culture are polyploid. Meaning there are two or more copies of the gene of interest due to whole chromosome duplication, or translocations. These alterations usually have occurred during disease progression, cell line immortalization, or continuous cell culture. To obtain a functional knockout in diploid or polyploid cell lines one would need to mutate every copy of the gene within the cells. A CRISPR knockout of each copy of these genes will have different mutations at the DNA level which can give different RNA transcripts and potentially different phenotypes in the cell.
Our HAP1 cell line is near-haploid meaning that they has, for the most part, one copy of a gene transcribed from a single allele. Since only one allele is expressed, only one allele needs to be knocked out (Figure 1). There are no additional copies of the gene which can mask the knockout or have a different phenotype making HAP1 cells a useful tool for studying gene function, eases the creation of these cells, and makes results easier to interpret.
What are HAP1 cells? Is my gene haploid in these cells?
The HAP1 cell line originated from KBM-7 (Catalog C628), a suspension cell line that was derived from cells taken from a myeloid leukemia patient (Andersson, B et al Cancer Genet. Cytogenet 1987, Kontecki et al 1999 Exp Cell Res). KBM-7 cells are near- haploid except for two copies of Chromosome (Chr) 8, and a small section of Chr 15 that is duplicated in Chr 19. As these cells are from a myeloid lineage, the KBM-7s have a classic marker of myeloid leukemia cells—the Philadelphia chromosome (Kotecki et al 1999 Exp Cell Res, Carette et al 2009 in Science).
| KBM-7 | HAP1 | E-HAP | |
| Catalog # | C628 | C631/C859* | C669 |
| Morphology | Suspension | Adherent | Adherent |
| Karyotype | Near-Haploid Except for Chr 8 and~300 gene duplication of Chr 15 in Chr 19 |
Near-Haploid Except for ~300 gene duplication of Chr 15 in Chr 19. |
Haploid |
| Philadelphia Chromosome | Present | Present | Present |
KBM-7 cells were transfected with Yamanaka factors (Oct4, cMyc, Sox2, and Klf4) to induce pluripotency. Although pluripotency was not achieved, cells transfected with the Yamanaka factors grew out a population of adherent fibroblast-like cells. The HAP1 cell line was obtained from clonal populations of these cells. HAP1 cells are different from KBM-7 cells in that the HAP1 cells are no longer diploid for Chr 8, but they retain the fragment of Chr 15 in Chr 19 (table 1). The fragment of Chr 15 that is duplicated is approximately 30 Mb and contains 330 genes. We use this HAP1 parental cell line to make our knockout cell lines.
What is the difference between the different parental (wildtype) HAP1 cell lines that are offered?
Horizon offers three different parental lines for the HAP1 cells. Our C631 is the free parental line included with purchase of any of our HAP1 knockout clones. It should be noted we have two additional parental cells that can be separately be purchased depending on your downstream assay. Horizon offers screening ready HAP1 cells (C859) which are the same as C631, but we have done some additional sorting on these cells to remove any cells that have diploidized spontaneously. In a quest to obtain a truly haploid model Horizon generated the E-Hap (C669) cell line by removing the duplication of chromosome 15 in chromosome 19 by using CRISPR-Cas9 (Carette et al., 2011). These cell line do still have the Philadelphia chromosome.(Table1)
Do HAP1 cells express my gene of interest? How come there is no knockout of my gene of interest?
HAP1 cells have been characterized by RNAseq analysis. This characterization provides an estimate expression of these gene in a HAP1 in basal, unstimulated conditions. On each HAP1 gene-specific product page we have included the Wildtype (WT) expression level in transcripts per million (TPM) from this analysis. We considers a gene with a TPM of 3 or less to not be expressed. Importantly, our expression data is for wildtype HAP1 cells, knockout cell lines expression profile data should be verified and characterized once the cell line is received.
In characterization of genes in HAP1, there is a set of genes that we deem highly essential and the genes that are diploid in these cell lines (Bruckstrummer et al. 2013). Knockout of these genes do not appear in our catalog. Additional characterization including of sequencing of HAP1 cells has been performed independently. (Essletzbichler et al., 2014:) If we do not have a knockout of your gene of interest, contacting your sales representative or Scientific Support team can put you in touch to see if we can produce this cell line as a custom project.
How are HAP1 knockout cells produced? And how do you ensure a knockout has been obtained?
Our HAP1 knockout cells are produced using CRISPR/Cas9 technology. In this process, a guide RNA (gRNA) forms a complex with Cas9 nuclease, and directs Cas9 to a specific locus, where it cleaves the DNA. The cut is repaired by non-homologous end joining (NHEJ). This repair often results in indels (small insertions and deletions) that can cause frameshift mutation or a premature stop codon in the gene. Cells from this editing are grown out clonally and then sequenced at the genomic DNA level to ensure a frameshift mutation is present. The disrupted sequence in these clones leads to a premature stop codon resulting in NMD of the transcript or a frameshift mutation resulting in a protein of a different composition. The gRNA used and the sanger sequencing result can be found on the certificate of analysis (CoA) of the HAP1 cell line on the product page. Both the gRNA and Cas9 are expressed transiently in the cells, so the cells can be best used as you see fit moving forward as these components will be lost from your final produced cell line. More about how these HAP1 knockouts are produced can be found in this video. Since every researcher’s assay is different, we recommend further characterization of the cell line upon receipt to see how the knockout cell is morphologically and functionally different to the wildtype.
Will the HAP1 cells diploidize?
HAP1 cells do spontaneously diploidize, and this is a normal part of their biology. It can normally be observed between passages 7- 10, with some HAP1 cell lines completely becoming diploid by passage 20. The knockout is introduced when the cells are haploid, so diploidization after this point will just result in the other allele having a copy of the knockout produced. Since this diploidization happens after the editing has taken place, the knockout status of the cell will not be affected by diploidization.
It is possible to isolate and propagate haploid HAP1s from the cell line pool. Since the HAP1 population is a mixed pool of haploid and diploid cells, the haploid HAP1s are roughly half the size of the diploid HAP1s. If a haploid population is desired or needed for a particular assay it is recommended to use the HAP1 cells from passages from 7 to 10 and do an enrichment of HAP1s by size by flow cytometry. Freezing down cells at an early passage will also make it easy to return to this haploid population. The literature has more about some of the characterized differences between the Haploid and diploid HAP1 cells (Beigl et al., 2020).
How can I differentially detect a knockout from the wildtype cell line?
CRISPR/Cas9 is used create our HAP1 knockouts, by introducing a premature stop codon or frameshift early in the gene. Some detection methods might fail to resolve the difference between the knockout and the parental cell line. The best way to validate your knock out is using a well-characterized functional assay that can delineate these differences between the mutant and parental cell line. Some examples of powerful ways to use HAP1 cells can be found in this Horizon Blog article “Beyond the western blot”.
Detecting mRNA by RT-PCR might reveal the transcript with the indel without regard to function. Validating the cell line with an antibody can also be challenging and misleading if the precise epitope of the antibody is not known and characterized. (See this article about validating your antibody). For a complete description of how the protein can be affected after knockout please visit this video.
What are the culturing requirements for HAP1?
HAP1 cells have specific culture requirements and should be cultured according to provided protocols. HAP1 cells should remain at or below 75 percent confluency. The doubling time is approximately 12-16 hours depending on precise culturing conditions. Note that, although HAP1 cells are adherent, we have observed them to ball up and detach from the plate when they duplicate their DNA in S-phase. When thawing cells or isolating single clones, performing half-media changes to ensure these cells are left behind and allowed to continue to divide. The HAP1 culturing protocol can be found here. In addition we have another blog post that will “set yourself up for success with HAP1 cells” that goes over additional points on maintaining these cells.
How can I use a HAP1 cell line in my study?
HAP1s are a powerful research tool. HAP1 knockouts can be important for antibody validation, or they can be used as proof of concept before creating a more complex knockout. If you have further questions about the HAP1 knockout cells feel free to reach out to Scientific Support by phone, chat, or at technical@horizondiscovery.com and we can discuss the potential to use HAP1s in your assay.
Find a HAP1 knockout cell line
Learn more about how HAP1 cells have been used
Contact Us
Featured Products
Browse our selection of HAP1 knockouts to find your gene of interest.
Buy a HAP1 parental cell line
Related Content
Why are there several HAP1 cell lines for my gene, how long will the take to be delivered Blog Article
Good experiments with HAP1 cells: parental and mutant cell lines, media controls, assay validation, ploidy Blog Article
Beyond the western blot and the advantages of HAP1 cells Blog Article
HAP1 cell references reading list Reading list
Top peer reviewed scientific articles using HAP1 cells Blog Article
HAP1 FAQs Blog Article
Application Two content goes here.
Additional Publications that describe HAP1 cell creation and characterization
- Andersson et al., Cancer and Genetics and Cytogenetics 1987. Ph-positive chronic myeloid leukemia with near-haploid conversion in vivo and establishment of a continuously growing cell line with similar cytogenetic pattern
- Kotecki, M., et al. Experimental Cell Research 252, 273–280 (1999). Isolation and Characterization of a Near-Haploid Human Cell Line.
- Essletzbichler P. et al., Genome Res. 2014. Genomic characterization of HAP1 cell line;
- Dong M. et al., Neurology 2014. HAP1 knockout cell line for evaluation of pathogenic mutations using phenotype rescue experiments;
- Kravtsova-Ivantsiv Y. et al., Cell 2015. HAP1 knockouts of KPC1 and KPC2 support role of KPC1 as E3 ligase that mediates processing of NF-kB1 p105 to p50
- Carette, J. E. et al. Science 2009. Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens.
- Carette, J. E. et al. Nature. 2011.Ebola virus entry requires the cholesterol transporter Niemann–Pick C1
- Lackner DH et al. Nat Commun. 2015. A generic strategy for CRISPR-Cas9-mediated gene tagging.
- Essletzbichler, Patrick et al. Genome research vol. 24,12 (2014) “Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line.”
- Beigl et al. Biology Open. 2020 Efficient and crucial quality control of HAP1 cell ploidy status.
- Burckstrummer et al. Nat Methods 2013 A reversible gene trap collection empowers haploid genetics in human cells