Overcoming the pitfalls of validating knockout cell lines by western blot
What happens to the protein after a frameshift mutation?
A frameshift mutation can result in a premature stop codon which is often presumed to lead to a knockout. Depending on the region targeted by the guide RNA, how the DNA is repaired by the cell, and the location of the stop codon1, there are multiple possible outcomes at the protein level:
- mRNA degradation by nonsense-mediated decay (NMD) before translation, so no protein is produced (Fig. 1b).
- translation to make a truncated protein (Fig. 1c).
- translation beginning from an alternative start codon after the premature stop codon producing a truncated protein (Fig. 1d).
- exon skipping, excluding the exon containing the indel, to produce a splice variant (Fig. 1e)
- translation of a truncated portion of the wildtype protein followed by a novel amino acid sequence (Fig. 1f).
Some of these fates can co-occur in the same cell line. For example, mRNA where the targeted exon is skipped may be present alongside mRNA including the targeted exon.2
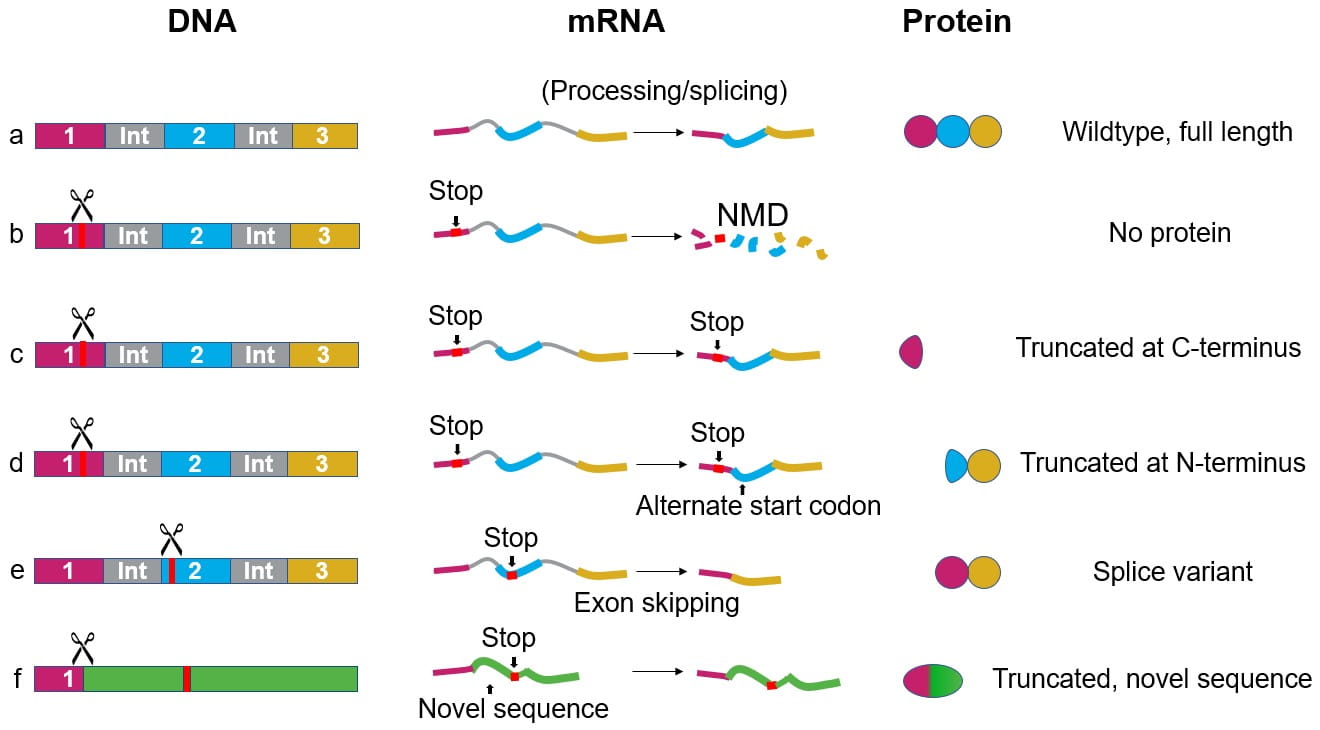 Figure 1. DNA, mRNA, and protein in a wildtype cell line (a) and possible outcomes after a frameshift mutation (b-f).
Figure 1. DNA, mRNA, and protein in a wildtype cell line (a) and possible outcomes after a frameshift mutation (b-f).
A common way to check clones for knockout is western blotting. Western blot results will depend on where the antibody binds, and the protein expressed by the cell line; figures 2 and 3 show examples of how a western blot could look.
Why are bands visible in a western blot using a knockout cell line?
The western blot in Figure 2 uses an antibody recognizing an epitope in the N-terminus of the wildtype protein. If the mRNA is destroyed by NMD, or if a splice variant without the N-terminus is generated, bands will not be observed on the blot. However, if the protein is truncated at the C-terminus, or if the wildtype N-terminus is followed by a novel amino acid sequence, a band may be observed. Such bands may lead to misinterpretation of the results.
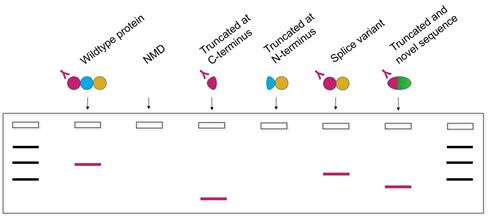 Figure 2. Schematic representation of a western blot showing protein bands detected by an antibody against the wildtype protein’s N-terminus, for each of the proteins outlined in Figure 1.
Figure 2. Schematic representation of a western blot showing protein bands detected by an antibody against the wildtype protein’s N-terminus, for each of the proteins outlined in Figure 1.
Using a second antibody, for example one binding an epitope in the center of the protein (Figure 3), will provide more information. Analyzing the bands observed in both blots gives additional insight into whether the wildtype protein is still present, but it cannot confirm whether the protein detected is functional.
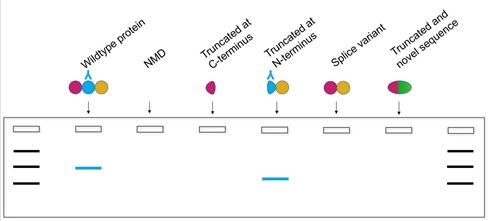
Therefore, we recommend using a functional assay specific to the protein of interest to assess a function which is important for downstream experiments. Here is a list of functional assays researchers have used with Horizon’s HAP1 knockout cell lines; this list of assays could be applied to check for protein function in any knockout cell line.
How can a knockout cell line be validated?
Whether the cell line in question is a HAP1 knockout cell line, a cancer-related knockout cell line, or a knockout cell line you have made yourself, here are some tips for validating the knockout:
- If possible, use a functional assay to investigate an important function of the targeted protein.
- In all assays, use the parental cell line as the control to account for cell line-specific effects.
- When using antibodies, ensure that they have been validated using positive and negative controls to ensure that they are binding the correct target.
- For all experiments, consider appropriate controls, including assay controls.
References
-
Popp, M. W. and Maquat, L. E., Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell 165 (2016). https://doi.org/10.1016/j.cell.2016.05.053
-
Tuladhar, R., Yeu, Y., Tyler Piazza, J. et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat Commun 10, 4056 (2019). https://doi.org/10.1038/s41467-019-12028-5
Other resources that you may be interested in:
• Antibody validation: Why is antibody validation important?
• Blog: Is your antibody binding the right target?
• Blog: Spying on proteins - how observation can affect the perception of reality
• Blog: Strategies to detect and validate your CRISPR gene edit
• Video: What happens to the protein in a knockout cell line?
