Improved functionality and specificity of genome editing experiments that only requires a gene search!
A big problem with current CRISPR-Cas9 tools for gene editing is that one of the two main required components, the guide RNA (which directs the Cas9 nuclease to cut at a specific genomic DNA target site), may cause a double strand break (DSB), but the DSB does not always cause functional knockout of the protein. Currently, researchers must use online design tools which rely on older, rudimentary rules with no basis for functionality, to design their guide RNA.
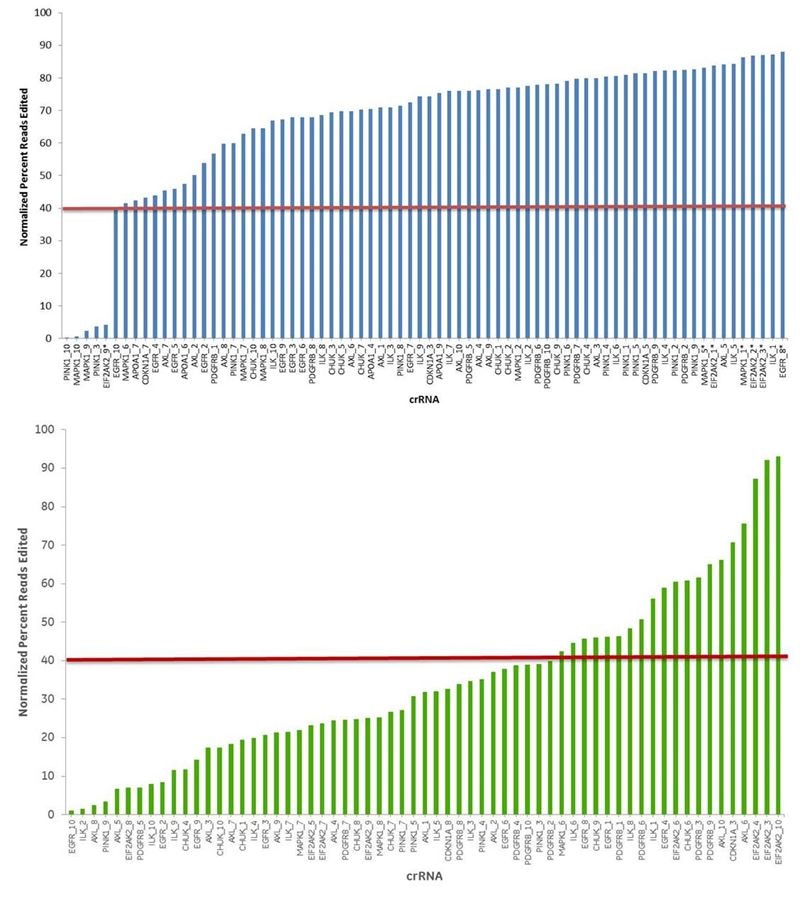
To address this issue, we developed the Dharmacon Edit-R CRISPR RNA algorithm, training it on over 1100 phenotypic data points to identify characteristics of truly functional guide RNAs. It has since demonstrated the ability to select highly functional guide RNAs across multiple assay systems.
 So we have an algorithm – what’s next?
So we have an algorithm – what’s next?
By leveraging our 20-year know-how in RNA chemistry and vector biology, we have released not one but TWO sets of genome-wide predesigned guide RNA products for human, mouse, and rat, all designed with our Edit-R algorithm. Now you only need to do a gene search, click, and order, to get either synthetic CRISPR RNA (crRNA) or lentiviral single guide RNA (sgRNA) products for functional target gene knockout.
To support every aspect of these experiments, we’ve also released extensive panels of positive and non-targeting controls and lentiviral sgRNA pooled screening libraries for broader studies across hundreds or thousands of genes.
Learn more
- Download our Open Access paper which demonstrates the importance of highly rigorous specificity-checking when designing CRISPR guide RNAs
- Learn how Cas9 expression levels impact resulting genome engineering, and how you can improve it by using the best promoter for your cells
- Review our poster on selection of optimal CRISPR-Cas9 reagents for improving experimental outcomes
