Cell free DNA formulated in synthetic matrix that is compatible with diverse extraction platforms provides greater flexibility in choosing reference materials.
What is liquid biopsy?

In cancer diagnostics, post-operative organ transplant monitoring, and prenatal genetic testing, one thing is common: early detection plays a pivotal role. While invasive biopsy procedures have historically been used to monitor relevant biomarkers of diseases, significant progress has been made over the last few years in non-invasive liquid biopsy techniques. Different types of body fluids can be used to perform liquid biopsy, with blood being the most common sample type for oncology. A simple blood draw has the potential for real time monitoring of treatment response and early cancer detection. Normal tissues and tumors shed cells or cell contents including DNA, and this circulating cell free DNA (cfDNA) released into peripheral blood is a widely used analyte in blood draws from cancer patients. Finding biomarkers using liquid biopsy is gaining traction and there are already a few FDA approved NGS-based companion diagnostic tests in use that monitor a range of solid tumors (i.e. Guardant360 Dx, FoundationOne Liquid CDx).
Cell-free DNA isolation and quality controls
Although fragmented DNA from both normal and tumor cells finds its way into the blood, the concentration of tumor specific cfDNA is very low. This low concentration and small fragment size mean that standard genomic DNA isolation techniques are not applicable for cfDNA isolation. Thus, choosing a robust cfDNA isolation process is a crucial step in determining the purity and yield of cfDNA which affects downstream analytical processes. Post cfDNA extraction, the analytical validity of a given test depends on the sensitivity and specificity of the assay.
To calibrate and validate the pre-analytical and analytical workflows, Horizon Discovery offers cfDNA reference standards that are formulated in synthetic matrix, which mimics human plasma, or buffer. These cell-line derived reference materials are commutable with patient samples, highly characterized and aid in assay development, validation and routine quality control in assays using cfDNA measurements (both qualitative and quantitative).
A varied number of extraction chemistries have been developed for isolating cfDNA from plasma, which largely involves silica-based purification columns and magnetic bead-based kits. Several studies have analyzed the efficacy of different extraction methods and have shown variabilities in yield and fragment sizes recovered (1-4). Other factors that can impact the DNA extraction process are physical conditions such as pH, temperature, and ionic strength. The presence of contaminating high molecular weight genomic DNA, proteins, metal ions and other common impurities in the cfDNA extract can also adversely affect certain molecular processes used in downstream analytical assays.
To combat the variability introduced by different extraction kits, Horizon Discovery has developed a new cfDNA reference standard in a synthetic matrix that is compatible with the most widely used extraction kits. Horizon Discovery now offers cfDNA reference material Synthetic Matrix II (HD917). This products differ in the composition of the artificial matrix formulation and therefore, they have varying levels of compatibility with the different extraction kits (Fig 1). This reference material offer more flexibility to customers in choosing their preferred control material based on their preference for a certain extraction kit.
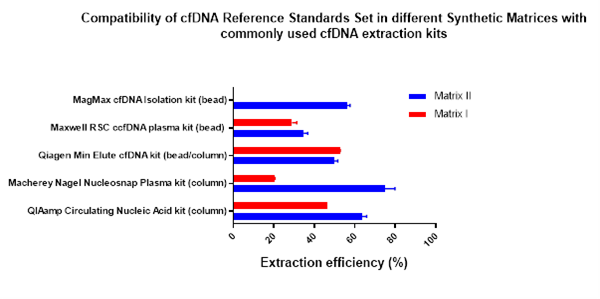
Analytical assessment of the reference material
One key challenge in early cancer detection and transplant rejection monitoring using cfDNA is the very low proportion of the mutated DNA or donor DNA in patients’ blood. The highly fragmented nature, very low concentration, and extremely low mutant allele fraction of cfDNA makes accurate detection of genetic variants substantially challenging and can limit clinical utility. Therefore, precise analysis of cfDNA requires use of appropriate controls with well characterized variants. Both, cfDNA reference material in Matrix I and II are offered at 5%, 1% and 0.1% mutant allele frequencies (MAF), in addition to a wild type isogenic product (0% allele frequency).
Key variants in the cfDNA reference standard are verified at multiple stages in the production process to ensure accuracy and consistency of the genetic composition, starting from the cell blending stage to the end, where fragmented cfDNA product is recovered from the artificial plasma. Digital PCR assays on the cfDNA fragments extracted from HD917 confirmed all the claimed variants at expected allele frequencies with high reproducibility (Fig 2).
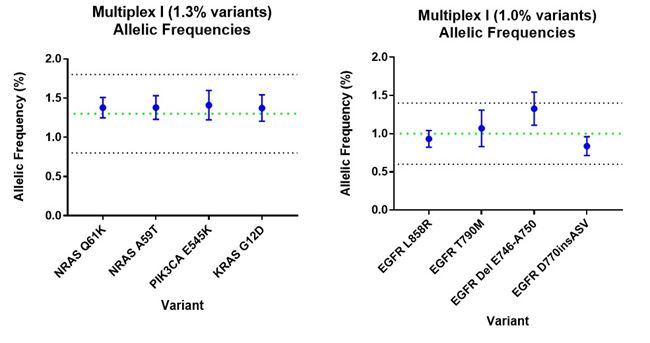 Whole exome sequencing of the cell blend used to make the reference standard also confirmed accurate detection of the claimed variants proving its utility across several diagnostic assays. In addition to variant verification, DNA fragment profile (170 bp average) and DNA concentration are also verified, meaning this reference standard can also be used to calibrate measurements of cfDNA physical parameters.
Whole exome sequencing of the cell blend used to make the reference standard also confirmed accurate detection of the claimed variants proving its utility across several diagnostic assays. In addition to variant verification, DNA fragment profile (170 bp average) and DNA concentration are also verified, meaning this reference standard can also be used to calibrate measurements of cfDNA physical parameters.
Summary
Horizon reference standards are cell line derived, which makes them an excellent renewable resource that mimics the complexities of the human genome. cfDNA reference material that is constructed using synthetic matrix is a cost-effective surrogate for human plasma, which suffers from lot-to-lot variability and requires special storage needs. With the availability of two different synthetic matrices and a range of compatibilities with cfDNA extraction kits, Horizon’s cfDNA reference standards are a great assay control for achieving confidence from start to finish in liquid biopsy assays.
 Written by Prabha Nagarajan
Written by Prabha Nagarajan
Dr. Nagarajan obtained her PhD from University of Rochester where she studied mitochondrial DNA repair mechanisms.
References
- Fong SL, Zhang JT, Lim CK, et al. Comparison of 7 methods for extracting cell-free DNA from serum samples of colorectal cancer patients. Clin Chem 2009;55:587-9. doi: 10.1373/clinchem.2008.110122
- Pérez-Barrios C, Nieto-Alcolado I, Torrente M, Jiménez-Sánchez C, Calvo V, Gutierrez-Sanz L, Palka M, Donoso-Navarro E, Provencio M, Romero A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: impact on biomarker testing. Transl Lung Cancer Res 2016;5(6):665-672. doi: 10.21037/ tlcr.2016.12.03
- Repiská G, Sedláčková T, Szemes T, et al. Selection of the optimal manual method of cell free fetal DNA isolation from maternal plasma. Clin Chem Lab Med 2013;51:1185-9.doi: 10.1515/cclm-2012-0418
- Holmberg RC, Gindlesperger A, Stokes T, et al. Akonni TruTip(®) and Qiagen(®) methods for extraction of fetal circulating DNA--evaluation by real-time and digital PCR doi: 10.1371/journal.pone.0073068
Interested in our liquid biopsy controls?
Want to learn more?
- Using reference standards in the development of new liquid biopsy platforms - blog
- Combatting the challenges of liquid biopsy with cfDNA synthetic plasma reference standards - blog
