250,000. That is the average number of data files (combination of genome, RNA, and protein) being added to the NCBI Reference Sequence Database (RefSeq) daily.1
As sequencing technologies continue to improve at astronomical rates, it now takes only a few hours to sequence an entire human genome – a feat that first took over 10 years and $2.7 billion to achieve. What does this mean for our understanding of the human genome and related transcriptome and proteome? It means it’s changing. Gaps are getting filled in. Complex regions are receiving more coverage. We’re getting better at understanding novel gene variants and where there are different isoforms occurring throughout the genome. What does that in turn mean for your research tools? It means they should be changing and adapting as well. That is why at the end of 2025 and moving forward into 2026, Revvity’s Dharmacon portfolio announced a major revamping, or what we’re calling, realignment, of our Edit-R portfolio of human synthetic and lentiviral predesigned guide RNAs. All of these guide RNAs were realigned to the most recent RefSeq in late 2025 to ensure they stay up to date with the constantly evolving genome for 2026 and beyond. Hence you will see a note such as this one below on many of our CRISPR guide RNA webpages:

Figure 1: Announcement of Edit-R sgRNAs updated to the latest RefSeq.
Whether you’re new to the Dharmacon portfolio of gene editing and modulation reagents, or a seasoned researcher who was using our first algorithmically derived siRNA reagents back in the early 2000s, this realignment should be important to you. Many CRISPR vendors develop an algorithm (often not empirically validated) and then design sgRNAs with that algorithm. However, an algorithm or algorithmically derived sgRNAs should only be a first step in that sgRNA design process. At Dharmacon, we’ve developed the Edit-R algorithm that has been extensively validated through a variety of functional assays and across thousands of designs, resulting in guide RNAs that efficiently generate functional protein knockouts, not just create insertions or deletions (indels). Our algorithm considers both functionality (ie: how likely is your guide to knockout your gene/protein of interest) and specificity (ie: how confident are you that your guide will not target elsewhere in the genome at off-target locations), both of which are very important considerations for guide performance and reliability. Other CRISPR reagent providers may also consider both functionality and specificity when developing an algorithm, but it is important to note that both functionality and specificity can be majorly compromised when guides are designed to a RefSeq that is not up-to-date. The now realigned Edit-R guides designed to the latest RefSeq ensures that the sgRNAs we provide continue to target intended genes with updated annotations. This, in turn, provides an added level of confidence to our customers and ensures that our guide RNAs are going to be the most scientifically up-to-date and biologically relevant for your research.
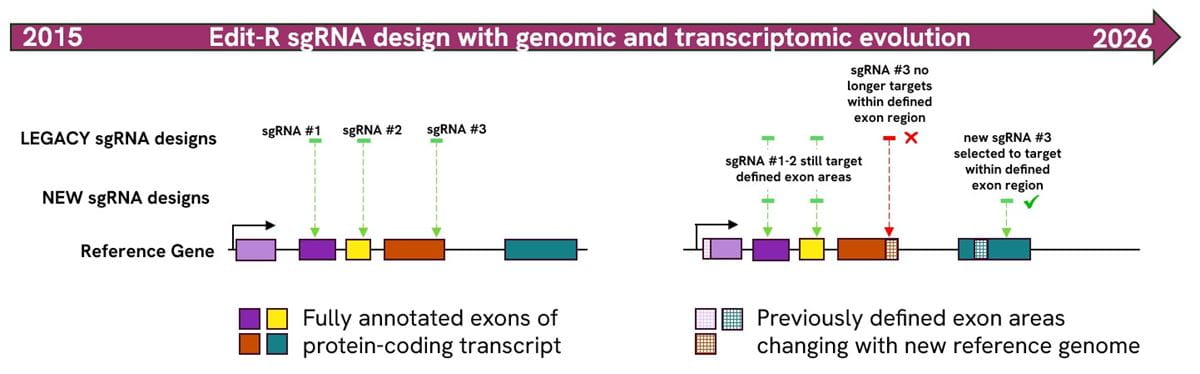
Figure 2: Schematic representation of Edit-R sgRNAs before and after realignment to the latest RefSeq. In this example, two of the three sgRNAs (Edit-R, set of 3) are the same before and after mapping to the updated reference gene. However, in this instance, sgRNA #3 no longer targets within a defined exon region so it has been replaced with a new sgRNA #3 that is predicted to efficiently induce a functional protein knockout with a high degree of specificity.
Below, we have generated a list of some of the most frequently asked questions following this realignment process.
Should I avoid using previously ordered guides that have been discontinued in favor of these new products?
Many guide RNAs that were replaced are still highly functional and specific; however, that gene target has at least one other, newer, guide RNA that now scores even higher and is more likely to induce a functional protein knockout while ensuring the smallest possible likelihood of targeting elsewhere across the genome. Additionally, guide RNAs for some gene targets remain unchanged before and after the realignment.
How do I know if a guide RNA that I ordered previously is still considered a top design for that gene?
Simply search for your catalog number at the top of our webpage. If you see the following message, it means that guide RNA was retired and replaced with one that has a higher combination of functionality and specificity scores when realigned to the latest RefSeq.
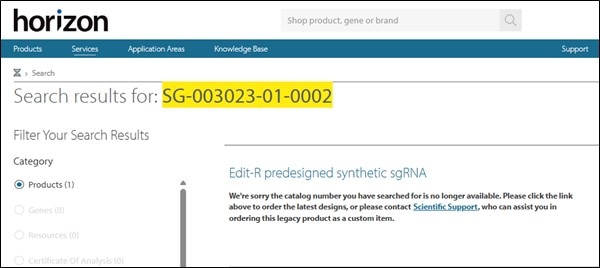
Figure 3: Searching by catalog number to identify if a guide RNA is active or retired.
What if a guide RNA that I worked with previously is no longer available but it works well in my experimental system?
Not a problem. That same guide RNA can still be ordered through our Guide RNA Designer tool that lets you input any 17-23 nt target sequence for automatic appending to the repeat region. Just enter in your targeting sequence and add to cart.
Why use NCBI RefSeq as your reference genome for this realignment instead of other reference resources such as Entrez or the UCSC Genome Browser? Can you please explain the differences?
NCBI RefSeq, Entrez, and UCSC Genome Browser all provide access to genomic sequence data and gene annotations, with Entrez serving as NCBI's search and retrieval system that links RefSeq sequences to literature (PubMed), protein structures, and other databases, while UCSC incorporates RefSeq data as one of its visualization tracks. The key differences are that RefSeq is a curated sequence database with standardized identifiers (NM_, NP_), Entrez is a federated search engine connecting multiple NCBI databases (including RefSeq, GenBank, PubMed, and protein databases) rather than a standalone genomic resource, and UCSC is an interactive visualization platform integrating RefSeq alongside other annotation sources (Ensembl, GENCODE, regulatory elements) for graphical genome exploration.
References:
- National Center for Biotechnology Information. About RefSeq. NCBI RefSeq. Retrieved 09/02/2025 from https://www.ncbi.nlm.nih.gov/refseq/about/
For additional questions, guidance, or support, please contact our technical support team at technical.horizon@revvity.com.
For additional information on how realignment is important for your research, see Part 1 of our Realigning and Remapping Dharmacon research tools series.
Further supporting your research:
In 2025, our Edit-R predesigned synthetic sgRNA prices were reduced by up to 50%. Effective January 1, 2026 an up to 30% price reduction was implemented on Edit-R predesigned All-in-one lentiviral sgRNA. Simply add to cart to see your new pricing or request a quote from your distributor.
