Why researchers are editing in T cells
Gene editing by CRISPR has led to significant advances in immunotherapy development. CRISPR can be used to rapidly knock out cell surface receptors, thereby limiting concerns of rejection by the immune system if the T cells engineered were not collected from the recipient. CRISPR also offers the ability to knock-in specific sequences into any gene, including chimeric antigen receptors (CARs) which target activated T cells to a specific tumor recognition site.
While the promise of using CRISPR to customize editing of immune cells is being realized, performing gene editing in this cell type poses many challenges compared to working with more robust, simpler to edit immortalized cell lines.
Tips for successful editing in T cells
Cell handling
T cells can be challenging to grow and maintain compared to immortalized cell lines.
- T cells are non-adherent; therefore, media changes and cell density monitoring will be more involved than when working with adherent cells.
- T cells can be manipulated in either an unstimulated or stimulated state. When editing in a stimulated state, the cells must be activated; and the optimal activation cocktail required for this will depend on the specific type of T cells being cultured, e.g., CD4s, CD8s, Tregs, etc.
Nucleofection
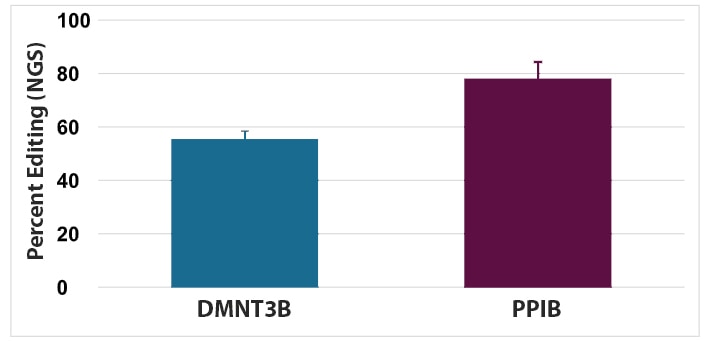
The optimal guide RNA and nuclease delivery conditions for T cells can exist in a narrow window, so extensive optimization is required to achieve reasonable editing efficiency.
- Conducting initial optimization work in an easier to manipulate surrogate cell line such as Jurkats (a leukemic T cell lymphoblast cell line) is recommended so one may be familiarized with the delivery and assay techniques.
- As there is donor-to-donor variability between different lots of collected human T cells, it is important to test and validate each new donor’s cells before proceeding with new experiments.
Analysis
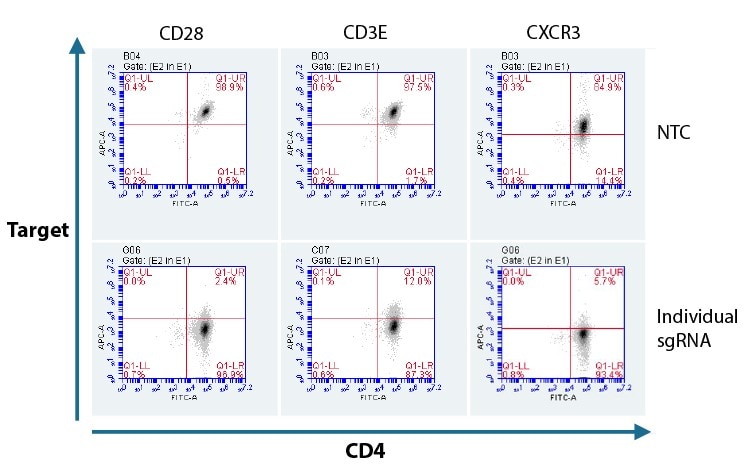
Flow cytometry is often the endpoint assay when working with T cells, as it allows researchers to study protein expression in live, intact cells. Assay optimization and validation, including finding the right antibodies and defining the staining parameters, will be required prior to assaying in edited T cells.
Solutions for editing T cells in your lab
The combination of Edit-R predesigned synthetic guide RNA and a DNA-free Cas9 nuclease (either mRNA or protein) achieves highly efficient gene editing and functional knockout in T cells, when delivered in optimal conditions following the tips above.
Search Edit-R synthetic sgRNA Order Cas9 mRNA Order Cas9 protein