The Horizon CRISPRmod CRISPR activation (CRISPRa) system is an adapted CRISPR-Cas9 system that is used for upregulation of genes. It utilizes a nuclease-deactivated S. pyogenes Cas9 (dCas9), that is fused to transcriptional activation domains (VP64, p65 and Rta). When paired with guide RNAs that target a gene near a promoter region, the gene's native transcription start site is activated.
There are several format options for dCas9-VPR. The dCas9-VPR mRNA allows for direct co-transfection or electroporation of reagents and provides options for enrichment by fluorescence or antibiotic selection. Lentiviral dCas9-VPR reagents can be used to generate a stable population of dCas9-VPR cells which is ideal for screening and for extended timepoint assays. Lentiviral dCas9-VPR reagents are available with three different promoters (hCMV, mCMV, or hEF1a) and can be supplied as either purified high-titer lentiviral particles or purified plasmid DNA.
CRISPRa for gene activation
CRISPR-Cas9 is not only for creating genomic double-strand breaks, it can also be used to target promoter regions to activate or inhibit transcription. Using a deactivated or dead Cas9 nuclease fused to transcription activators, like Cas9-VPR, in combination with a guide RNA in either a synthetic sgRNA or crRNA or lentiviral format, the endogenous expression of a gene can be up-regulated by anywhere from 5 to 50,000+ fold!
dCas9-VPR reagents for optimization and enrichment
Enrichment for gene activation can now easily be done using the dCas9-VPR mRNA reagents that co-express either EGFP or puromycin with dCas9-VPR. These reagents allow easy co-transfection or electroporation for optimization and visualization of successful delivery.
Choose from a variety of dCas9-VPR vectors
Promoter activity will vary by cell type and will affect dCas9-VPR expression, choosing an optimal promoter is important for robust gene overexpression. Lentiviral dCas9-VPR reagents are available with three different promoters (hCMV, mCMV, or hEF1a) and can be supplied as either purified high-titer lentiviral particles or purified plasmid DNA.
Table 1. SMARTchoice promoter options for expressing dCas9-VPR nuclease| Promoter | Description |
|---|---|
| hCMV | human cytomegalovirus immediate early promoter |
| mCMV | mouse cytomegalovirus immediate early promoter |
| hEF1α | human elongation factor 1 alpha promoter |
dCas9-VPR mRNA
-
CRISPRa dCas9-VPR mRNA
Purified dCas9-VPR mRNA for transient expression. Fluorescent and puromycin options available for sorting, enrichment, and visualization of delivery.
Vector-based dCas9-VPR
-
New CRISPRa dCas9-VPR expressing stable cell lines
Choose from a variety of popular cell backgrounds, ready for you to deliver a CRISPRa guide RNA for gene overexpression studies.
-
CRISPRa dCas9-VPR expression reagents
Generate your own stable dCas9-VPR expressing cells with purified lentiviral particles or plasmid DNA reagents.
Fold activation by CRISPRa varies by gene and depends on the endogenous gene expression level
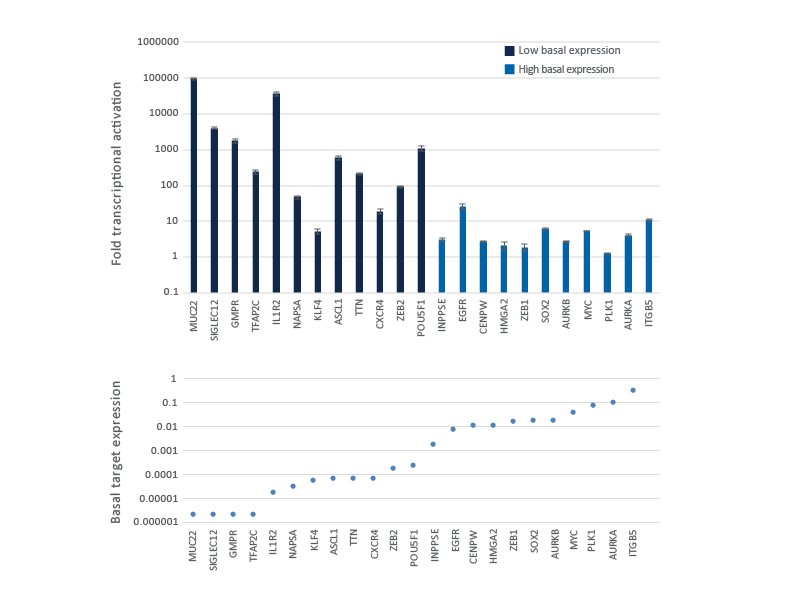
U2OS cells stably expressing integrated CRISPRa dCas9-VPR were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA pools (25 nM) targeting genes with low to high basal transcript expression levels. Cells were harvested 72 hours post-transfection and the relative gene expression was calculated using qRT-PCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control. The fold activation is shown for the genes ranked from low to high basal transcript expression level in samples treated with NTC control and is shown in the lower graph as basal gene expression relative to GAPDH expression in the same samples.
CRISPRa gene activation in U2OS cells is observed at 24 hours and increases at 48-72 hours

U2OS cells stably expressing integrated CRISPRa dCas9-VPR were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA targeting EGFR, IL1R2, POU5F1 or TFAP2C. Four CRISPRa crRNAs were used either individually or pooled (to a total concentration of 25 nM). Cells were harvested at 24, 48, and 72 hours post-transfection and the relative gene expression was calculated using qRT-PCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
CRISPRa gene activation by electroporation of dCas9-VPR mRNA and synthetic guide RNA

THP-1 and K-562 cells were electroporated using the Lonza 2b system with either CRISPRa dCas9-VPR mRNA (5 µg, Cat #CAS12024), CRISPRa Puro dCas9-VPR mRNA (5 ug, Cat #CAS12026) or CRISPRa EGFP dCas9-VPR mRNA (5 ug, Cat #CAS12025), CRISPRa synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting TTN (5 uM, Cat #P-005395-01-0005) or non-targeting control (NTC, 25 nM, Cat #U-009500-10-05). Cells were harvested at 48 hours post-transfection and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
Puromycin can be used to select for cells with CRISPRa Puro dCas9-VPR mRNA and enrich for gene activation

U2OS cells were plated at 200,000 cells per well in clear 6-well plates. After 24 hours, cells were co-transfected with CRISPRa Puro dCas9-VPR mRNA (2 µg, Cat #CAS12026), CRISPRa synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting either POU5F1 (25 nM Cat #P-019591-01-0005), IL1R2 (25 nM, Cat #P-007960-01-0005), TFAP2C (25 nM, Cat #P-005238-01-0005), TTN (25 nM, Cat #P-005395-01-0005) or non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (2 µg/well, Cat #T-2010-01). At 24 hours post-transfection, 2 µg/ml of puromycin growth media was added to the cells and a duplicate plate received normal growth media. At 48 hours post-transfection a crystal violet assay was performed and images captured to assess viability and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control. For all genes tested, we observed 3- to 5-fold enrichment of activation when compared to unselected samples.
EGFP can be used to select for cells with CRISPRa EGFP dCas9-VPR mRNA and enrich for gene activation
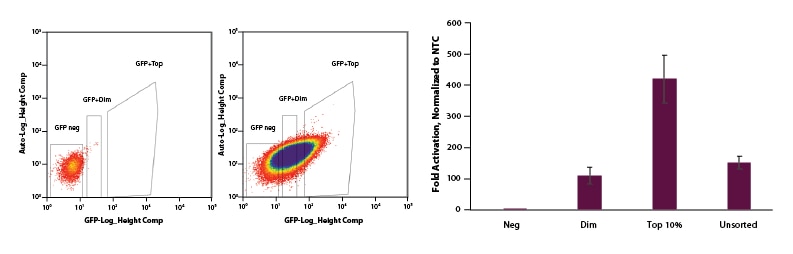
U2OS cells were plated at 200,000 cells per well in clear 6-well plates. After 24 hours, cells were co-transfected with CRISPRa EGFP dCas9-VPR mRNA (2 µg, Cat #CAS12025), CRISPRa synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting IL1R2 (25 nM, Cat #P-007960-01-0005) or Non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (2 µg/well, Cat #T-2010-01). At 24 hours post-transfection, cells were trypsinized and FACS was performed and cells were sorted into three categories: Negative, Dim, and Top 10%. Cells were replated in 6-well dishes and allowed to recover. After 24 hours, total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control. The negative population of cells sorted shows no activation. The Dim and Unsorted populations both showed ~120-fold activation and our Top 10% population resulted in 450-fold activation.
Efficient gene activation with dCas9-VPR mRNA and target sgRNA
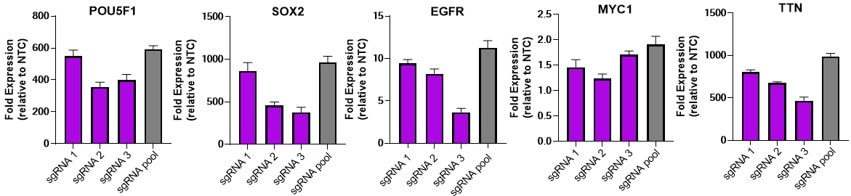
U2OS were plated at 10,000 cells/well and transfected using DharmaFECT Duo (0.15uL/well) Transfection Reagent with 25nM individual or pooled CRISPRa synthetic single guide RNA (sgRNA) targeting POU5F1, SOX2, EGFR, MYC1 or TTN and 200 ng / well of CRISPRmod CRISPRa dCas9-VPR mRNA. Cells were harvested at 72 hours post-transfection and gene expression was assessed using RT-qPCR. Relative fold transcriptional activation for each gene was calculated with the Cq method using beta-actin as the housekeeping gene and normalized to an experiment using non-targeting control sgRNA.
