- Gene editing
- Gene editing reagents
- Edit-R tracrRNA
Edit-R tracrRNA
Chemically synthesized trans-activating CRISPR RNA required for use with synthetic crRNA for fast and easy gene editing.

Please note our tracrRNA catalog numbers have changed to reflect the addition of chemical modifications to resist nuclease degradation. Modified tracrRNA is suitable for the same applications as the unmodified tracrRNA (U-002000-*) and can be used with either modified or unmodified Edit-R crRNA. These stabilizing modifications are required only for co-electroporation with Cas9 mRNA. Learn more about these changes in this featured article.
The Edit-R tracrRNA is a chemically synthesized and HPLC-purified long RNA molecule based on the optimized published S. pyogenes tracrRNA sequence (Jinek, 2012). It is chemically modified for nuclease resistance to improve performance in applications that use co-delivery with DNA-free Cas9 nuclease. The Edit-R tracrRNA is required for use with synthetic Edit-R crRNA, comprised of 20 nucleotides identical to the genomic DNA target site, or protospacer, followed by the required S. pyogenes repeat sequence that interacts with the tracrRNA.
Once delivered to the cell, the crRNA:tracrRNA complex with Cas9 nuclease to generate site-specific, DNA double-strand breaks (DSBs). When DSBs are repaired through non-homologous end-joining (NHEJ), the resulting small insertions and deletions (indels) can cause nonsense mutations resulting in gene disruption to produce a functional knockout.
Highlights
The Dharmacon Edit-R CRISPR-Cas9 synthetic crRNA platform requires three components for gene editing in mammalian cells, based on the natural S. pyogenes system:
- A protein, mRNA, or lentiviral vector expressing a mammalian codon-optimized gene sequence encoding Cas9 nuclease
- A chemically synthesized trans-activating CRISPR RNA (tracrRNA), and
- A chemically synthesized CRISPR RNA (crRNA) designed to the gene target site of interest
How much crRNA & tracrRNA do I need?
This table provides the approximate number of experiments that can be carried out for lipid transfection methods at the recommended crRNA:tracrRNA working concentration (25 nM:25nM) in various plate/well formats. Calculations do not account for pipetting errors.
For crRNA libraries, use (# wells)*(nmol per well) to determine the approximate amount of tracrRNA required. For instance, a 100 well library at 0.5 nmol per well would require 50 nmol of tracrRNA. The bulk sizes of tracrRNA are recommended for large library projects.| crRNA nmol | tracrRNA nmol | 96-well plate 100 µL reaction volume | 24-well plate 500 µL reaction volume | 12-well plate 1000 µL reaction volume | 6-well plate 2500 µL reaction volume |
|---|---|---|---|---|---|
| 2 | 2 | 800 | 160 | 80 | 32 |
| 5 | 5 | 2000 | 400 | 200 | 80 |
| 10 | 10 | 4000 | 800 | 400 | 160 |
| 20 | 20 | 8000 | 1600 | 800 | 320 |
Cas9 nuclease is programmed by crRNA:tracrRNA to specifically cleave target DNA
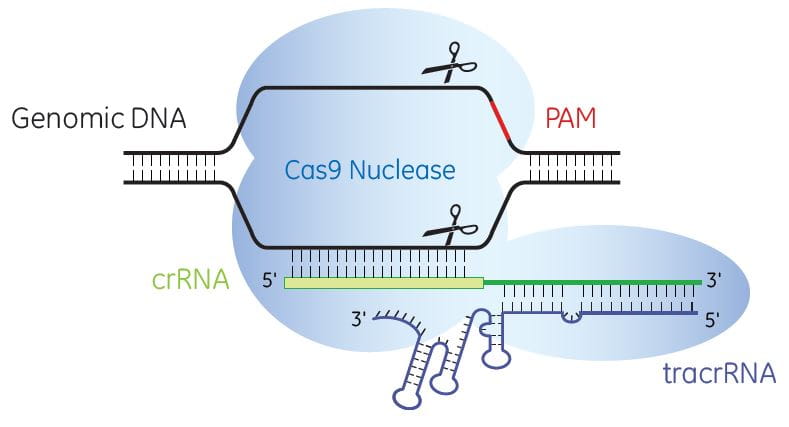
Once delivered to the cell, the crRNA and tracrRNA complex with Cas9 nuclease to generate site-specific, double-strand DNA breaks (DSBs). When DNA DSBs are repaired through non-homologous end-joining (NHEJ), the resulting small insertions and deletions (indels) can cause nonsense mutations and truncation of protein products or the introduction of a stop codon to produce gene knockouts.
UPLC trace demonstrating excellent purity of Dharmacon Edit-R synthetic tracrRNA
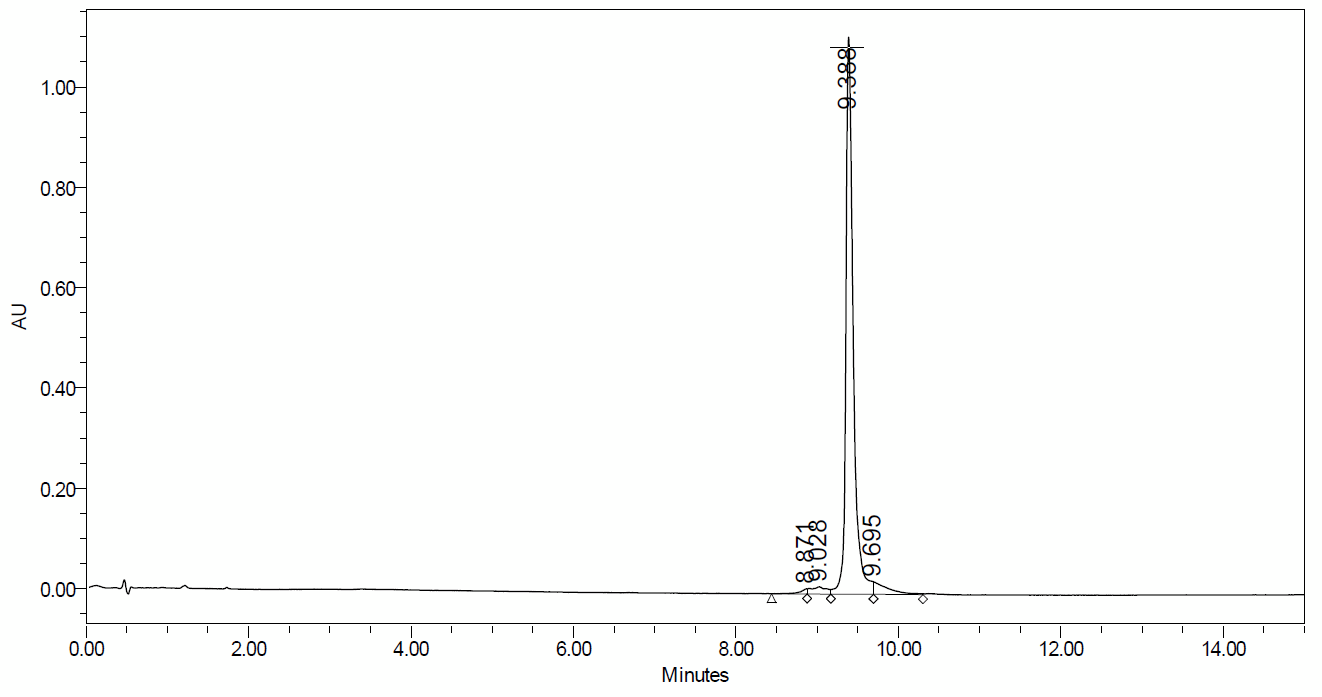
UPLC (ultra-performance liquid chromatography) analysis of Edit-R tracrRNA, a >70nt purified RNA, demonstrates the high quality achieved routinely by Dharmacon chemical synthesis. CRISPR-Cas9 experiments using a crRNA:tracrRNA to recruit Cas9 nuclease rely on excellent purity and sequence fidelity for optimal activity. Instrument used: Acquity UPLC by Waters.
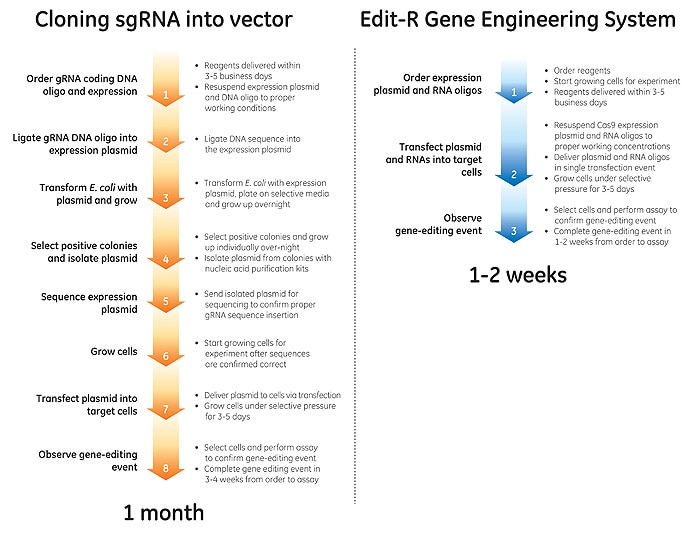
Less time preparing, more time experimenting
- M. Jinek, K. Chylinski, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 337(6096), 816-821 (2012).
-
M.L. Kelley, M.L., Ž. Strezoska, et al. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J. Biotechnol. 233, 74–83 (2016). doi:10.1016/j.jbiotec.2016.06.011
Application notes
-
A CRISPR-Cas9 gene engineering workflow: generating functional knockouts using Edit-R Cas9 and synthetic crRNA and tracrRNA - Application Note
-
Homology-directed repair with Dharmacon Edit-R CRISPR-Cas9 reagents and single-strand DNA oligos - Application Note
-
Optimization of reverse transfection of Edit-R synthetic crRNA and tracrRNA components with DharmaFECT transfection reagent in a Cas9-expressing cell line - Application Note
-
Using the CRISPR-Cas9 system with paired Dharmacon Edit-R synthetic crRNAs for functional knockout of microRNA hsa-miR-221 - Application Note
Posters
-
2'-O-methyl phosphorothioate linkage-modified synthetic guide RNAs for efficient CRISPR-Cas9 genome editing and reduced cellular toxicity - Poster
-
A Synthetic CRISPR-Cas9 System for Homology-Directed Repair - Poster
-
An algorithm for selecting highly functional and specific guide RNAs for CRISPR-Cas9 gene knockout - Poster
-
Complete alignment identification of CRISPR-Cas9 genomic off-targets using Edit-R CRISPR specificity tool - Poster
-
CRISPR-Cas9 genome editing utilizing chemically synthesized RNA - Poster
-
Homology-directed repair with Edit-R CRISPR-Cas9 and single-strand DNA oligos - Poster
-
Increasing gene editing efficiencies in eukaryotic cell lines by selection of appropriate CRISPR-Cas9 reagents - Poster
Product data
Product inserts
Protocols
Quick protocols
Safety data sheets
Related Products
Synthetic guide RNAs for efficient gene knockout and unparalleled specificity
Predesigned synthetic guide RNA for over-expression of human and mouse genes using CRISPR activation. Just search for your gene! Available as pooled or individual reagents.
Perform rapid functional enhancement screening with arrayed CRISPR activation guide RNA libraries. Find predefined collections for pathway, druggable, gene family or whole genome screening
