Ensure your system is optimized for successful CRISPR-Cas9 experiments
Edit-R synthetic sgRNA
-
Positive controls and detection primers
Species-specific sgRNAs targeting well-characterized genes, as well as mismatch detection assay primers, to determine the effectiveness of your gene editing conditions for maximal efficiency.
-
Non-targeting controls
Non-targeting controls to evaluate cellular responses to CRISPR-Cas9 components in the absence of gene target-specific sgRNA.
-
Cutting controls
Cutting control (safe harbor) synthetic sgRNAs recommended for determination of baseline cellular responses in CRISPR-Cas9 experiments.
Edit-R synthetic crRNA
-
Positive controls and detection primers
Species-specific crRNAs targeting well-characterized genes, as well as mismatch detection assay primers, to determine the effectiveness of your gene editing conditions for maximal efficiency.
-
Non-targeting controls
Non-targeting controls to evaluate cellular responses to CRISPR-Cas9 components in the absence of gene target-specific crRNA.
-
Cutting controls
Cutting control (safe harbor) crRNAs recommended for determination of baseline cellular responses in CRISPR-Cas9 experiments.
Edit-R all-in-one lentiviral sgRNA
-
Positive controls and detection primers
Control all-in-one lentiviral sgRNAs to verify DNA double-strand breaks and gene editing efficiencies.
-
Non-targeting controls
All-in-one lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human, mouse or rat genomes.
-
Cutting Controls
Cutting control (safe harbor) sgRNA recommended for determination of baseline cellular responses in CRISPR-Cas9 experiments.
Edit-R lentiviral sgRNA
-
Positive controls and detection primers
Species-specific sgRNAs targeting well-characterized genes to determine the effectiveness of your gene editing conditions for maximal efficiency.
-
Non-targeting controls
Non-targeting controls to evaluate cellular responses to CRISPR-Cas9 components in the absence of gene target-specific sgRNA
-
Cutting Controls
Cutting control (safe harbor) sgRNA recommended for determination of baseline cellular responses in CRISPR-Cas9 experiments.
The importance of effective controls in CRISPR-Cas9 experiments
The CRISPR-Cas9 system requires multiple components to be delivered to the cell for effective gene knockout, both a guide RNA and a Cas9 nuclease. Insufficient amounts of either component will result in poor gene editing.
Use of a species-specific positive control with our validated detection primers in a mismatch assay will verify the suitability of your experimental conditions for good CRISPR-Cas9 gene editing, and serve as an indicator of good transfection in ongoing experiments.
Our panel of Non-targeting controls permits assessment of potential non-specific effects, and lets you choose the optimal negative control for your cell type, and your assay.
DNA mismatch detection assays (SURVEYOR® or T7EI) using our detection primers are a relatively fast and high-throughput method for assessing gene knockout results in clonal or mixed cell populations. While they can only assess the relative percentage of insertions and deletions (not functional protein knockout), they are highly useful for:
- Optimizing crRNA:tracrRNA transfection conditions
- Determining the optimal promoter for driving Cas9 nuclease expression
Supporting data
Effective gene editing of PPIB in human and mouse cells
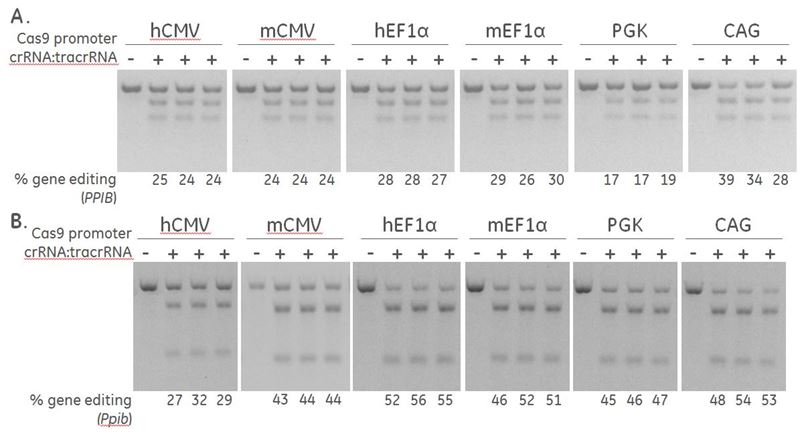
Positive control crRNA and Detection Primers targeting PPIB are used to determine the optimal promoter for driving expression of Cas9 nuclease. A human recombinant U2OS ubiquitin-EGFP proteasome cell line (Ubi[G76V]-EGFP) (A) and a mouse fibroblast (NIH/3T3) (B), were stably transduced with lentiviral particles containing Cas9 and a blasticidin resistance gene driven by the indicated promoters. A population of cells with stably integrated Cas9-blastR was selected with blasticidin for a minimum of 10 days before transfections. Cells were transfected with 50 nM synthetic crRNA:tracrRNA targeting Human PPIB / mouse Ppib using DharmaFECT 1 and DharmaFECT 3 Transfection reagent, respectively. After 72 hours, the relative frequency of gene editing was calculated based on a DNA mismatch detection assay using T7EI on genomic DNA extracted from the transfected cells.
