Enzymatically synthesized guide RNA can trigger unwanted cellular responses
Cellular delivery of both guide RNA and Cas9 nuclease is required for gene editing with the CRISPR-Cas9 system. Depending on the type of guide RNA used, this exogenous RNA can cause unwanted effects, such as an immune response, and interfere with experimental observations. The mammalian innate immune system recognizes whether RNA is from itself or foreign invaders (bacteria, fungi, viruses). If a cell identifies RNA as foreign, it mounts an immune response that can result in cell death. Different immune response pathway proteins are responsible for recognition of RNA characteristics: single-stranded RNA, double-stranded RNA, triphosphates on the end of single-stranded RNA, and even the length of the RNA.1,2
Guide RNA can be enzymatically or chemically synthesized
In vitro transcribed single guide RNA (sgRNA) is generated enzymatically using a DNA template, ribonucleoside triphosphates and a bacteriophage RNA polymerase, most commonly T7.3 After transcription, a 5’ triphosphate remains on the RNA which requires removal by a phosphatase enzyme. Finally, RNA must be purified from unincorporated triphosphates, protein and DNA. While labor-intensive, enzymatic synthesis can be performed in most laboratories and is cost-effective.4
Both crRNA:tracrRNA and sgRNA can be generated by chemical synthesis, which builds RNA using nucleoside phosphoramidites on a solid support. This is followed by cleavage from the support, deprotection, and desalting to remove impurities (reviewed in Kelley, 2016)5. Automated solid-phase RNA synthesis can generate RNA of different lengths and sequences, and it enables the addition of chemical modifications in specific locations.
The innate immune response to different guide RNA formats
To measure whether different formats of guide RNA cause unwanted effects, we performed an experiment where editing efficiency, viability, and up-regulation of immune response genes were monitored following delivery for several types of guide RNA (Table 1). For the chemically synthesized guide RNAs, we utilized similar chemical modification approaches to those described previously for siRNAs, antisense RNAs, and guide RNAs6-8. Chemical modifications included phosphorothioate on the RNA backbone and 2’-O-methyl on the sugar to improve the nuclease stability of the guide RNA.
|
Format of guide RNA tested |
Chemical modifications |
Generated by: |
|---|---|---|
|
crRNA: tracrRNA |
|
Chemical synthesis |
|
Single guide RNA (sgRNA) |
|
Chemical synthesis |
|
In vitro transcribed sgRNA (IVT) |
None |
Enzymatic synthesis (in vitro transcription) |
While all the guide RNA formats showed comparable editing by mismatch detection assay5, some of the guide RNAs dramatically affected the immune response genes (Figure 1). Notably, in vitro transcribed sgRNA induced immune response genes to similar levels induced by the positive control (poly I:C). Synthetic sgRNA and crRNA:tracrRNA without modifications had no detectable immune response. Synthetic sgRNA and crRNA:tracrRNA with two phosphorothioate linkages and two 2’-O-methyl (2xMS) modifications on one end (crRNA:tracrRNA) or both 5’ and 3’ ends (sgRNA) also had no detectable immune response. However the crRNA:tracRNA with 3xMS modifications on both 5’ and 3’ ends had a moderate immune gene induction and some cell death, and sgRNA with 3xMS 5’ and 3’ ends modifications had one instance (DNMT3B, OAS2) where immune response gene’s expression was induced.
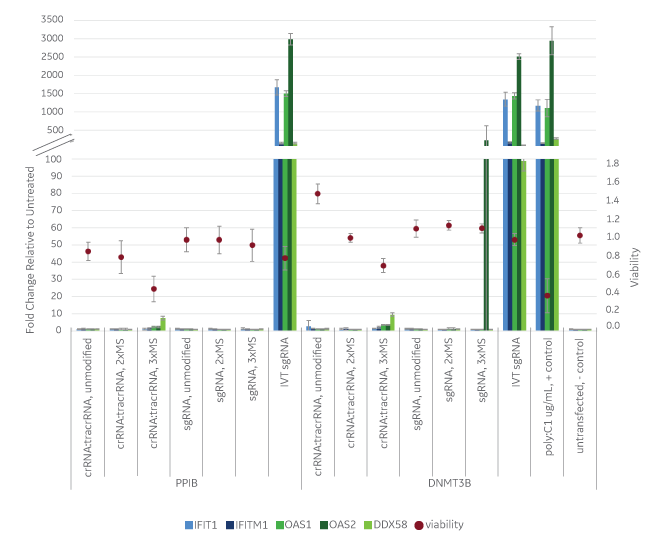
Edit-R synthetic guide RNAs cause virtually no innate immune response or toxicity compared to in vitro transcribed guide RNA
Judicious modification of synthetic guide RNA ensures good function without unwanted toxicity
These results are consistent with previous observations in the RNA field where cell death and an activation of the immune response can occur in some cell types due to incomplete removal of 5’ triphosphates from an in vitrotranscribed RNA9 or due to remaining impurities from the reactions. For the modifications on the chemically synthesized RNAs, it is known that some modifications can actually help RNA evade the innate immune system while other modifications can lead to cellular toxicity.6,8 Therefore, it is important to use modifications that improve characteristics of the molecule while not inducing responses that can complicate experimental outcome and interfere with phenotypic observations. Both Edit-R crRNA:tracrRNA and Edit-R sgRNA utilize chemical modifications that ensure good function (via increased nuclease resistance in some delivery modalities) without unwanted effects.
Authors: Emily M. Anderson and Annaleen Vermeulen, Senior Scientists at Dharmacon
References
- Reynolds et al., Induction of the interferon response by siRNA is cell type- and duplex length-dependent, RNA 12(6):988-93 (2006), 10.1261/rna.2340906
- Judge et al., Overcoming the Innate Immune Response to Small Interfering RNA, Human Gene Therapy 19(2): 111-124 (2008), doi: 10.1089/hum.2007.179
- Cho et al., Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease, Nature Biotechnology 31, 230–232 (2013), doi:10.1038/nbt.2507
- Edelmann et al., Production of pure and functional RNA for in vitro reconstitution experiments, Methods 65 (3) 333–341 (2014), doi: 10.1016/j.ymeth.2013.08.034
- Kelley et al., Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing, Journal of Biotechnoly, 10;233:74-83 (2016), doi: 10.1016/j.jbiotec.2016.06.011
- Dirin et al., Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides, Expert Opinion on Biological Therapy, 13:6, 875-888 (2013), doi: 10.1517/14712598.2013.774366
- Hendel et al., Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells, Nature Biotechnol., 33, 985–989 (2015), doi:10.1038/nbt.3290
- Dowdy, Overcoming cellular barriers for RNA therapeutics, Nature Biotechnology 35, 222–229 (2017), doi:10.1038/nbt.3802
- Kim et al., Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase, Nature Biotechnology 22, 321 - 325 (2004), doi:10.1038/nbt940
Products
-
Edit-R predesigned synthetic sgRNA
Guaranteed to edit your human gene target! Algorithm-optimized to maximize functional knockout and minimize off-target effects. Simply search for your gene! -
Edit-R synthetic sgRNA screening libraries
Arrayed collections of pooled pre-designed synthetic sgRNA for screening across the human genome and gene families. -
Edit-R predesigned synthetic crRNA
Algorithm-optimized crRNA for genome-wide coverage of human, mouse, or rat genes.
Additional Resources
-
CRISPR-Cas9 Gene Editing - Applications
CRISPR-Cas9 systems can be used with custom RNA guides for several gene editing applications
