Verify proper intracellular localization from HDR experiments with an IN Cell Analyzer 2200
Insertion of a fluorescent reporter using homology-directed repair (HDR) is often met with frustration due to inefficient knock-in rates and a significant workload to screen and identify the clonal cell line that contains the intended genomic modification.
The solution
Efficient gene editing reagents for knock-in and the appropriate equipment for observing fluorescent cells make it significantly easier to identify cells with the correct and functional insert.
Resolution matters when verifying intracellular localization of TurboGFP
We used Dharmacon Edit-R reagents to generate a TurboGFP N-terminal fusion to the ACTB gene in U2OS cells. We passaged the cells twice before visualization on two different epifluorescent, widefield microscopes seven days post-transfection. We tried to determine correct TurboGFP-ACTB localization on the same HDR cell population with an older microscope (which will remain unnamed) and an IN Cell Analyzer 2200. While the older microscope had sufficient magnification to visualize the cells when using a 40x objective, the green fluorescence was not at all sharp and we could not accurately determine correct ACTB localization, neither with visual inspection nor with ImageJ analysis software (Figure 1). However, upon using the IN Cell Analyzer with only a 20x objective, we were able to very clearly visualize TurboGFP-labeled actin filaments in the cytoplasm. The age of the microscope, CCD camera, and filter cubes all likely contribute to the older microscope’s poor quality images, even when used at higher magnification. Using the latest generation of fluorescent microscopes, such as the IN Cell Analyzer 2200, will greatly aid in evaluating correct insertion of the fluorescent tag while maintaining correct localization and function.
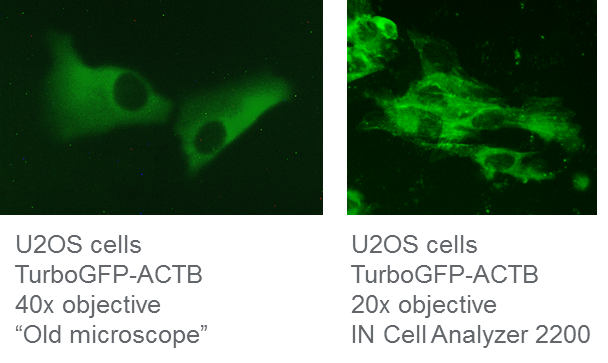
IN Cell Analyzer 2200 improves visualization of HDR-mediated fluorescent knock-in
CRISPR-Cas9 HDR experimental summary
Briefly, here is how we created a TurboGFP N-terminal fusion to the ACTB gene. U2OS cells were transfected with 50 nM ACTB-crRNA:tracrRNA, 25 nM Cas9 protein, 200 ng/µL ACTB TurboGFP HDR donor plasmid, and 0.3 µL/well DharmaFECT Duo transfection reagent and visualized on different fluorescent microscopes seven days post-transfection.
The crRNA was designed adjacent to the N-ternimus of the ACTB gene using the CRISPR Design Tool using the HDR settings.
Author: John A Schiel, R&D Scientist at Dharmacon
Additional Resources
HDR Donor Designer
- Design and order a custom donor oligo or plasmid for HDR-mediated gene editing in over 30 different species
Edit-R HDR Plasmid Donor Kits
- Rapidly and easily assemble a plasmid donor for HDR
