Now available: Whole exome sequencing (WES) data for Horizon’s most popular products! We now provide high quality NGS data sets (very high read depth of >500x), enabling detection of false positives and confirmation of true positives in your own data sets.
Here we explore what you can expect to find in the VCF files and how this can help build confidence in your applications.
We listened to feedback that our exome sequenced OncoSpan product data is highly valuable for analysis pipelines and have expanded our exome data offering to include a number of other NGS products in both gDNA and cfDNA formats.
What products were sequenced?
| Catalog number | Description | Batch number sequenced | Source material |
|---|---|---|---|
| HD728 | Tru-Q1 | 40707 | gDNA |
| HD729 | Tru-Q2 | 40755 | gDNA |
| HD730 | Tru-Q3 | 40780 | gDNA |
| HD731 | Tru-Q4 | 40833 | gDNA |
| HD734 | Tru-Q7 | 40919 | gDNA |
| HD752 | Tru-Q0 | 43262 | gDNA |
| HD753 | Structural Multiplex gDNA | 41130 | gDNA |
| HD776 | Multiplex cfDNA 0% | 46125 | gDNA |
| HD777 | Multiplex cfDNA 5% | 46125 | gDNA |
| HD793 | BRCA Germline | 46024 | gDNA |
| HD795 | BRCA Somatic Multiplex I | 46500 | gDNA |
| HD802 | EGFR Quantitative Multiplex | 46026 | gDNA |
| HD829 | Myeloid gDNA | 47035 | gDNA |
Table 1. All sequenced Horizon products with relevant information.
How were the products sequenced?
All samples were sequenced at the Genomics facility in Pittsburgh, PA, USA. The kits used in the sequencing workflows are shown in Table 2 below.
| Product type | Library generation kit | WES probe | Capture kit | Sequencer |
|---|---|---|---|---|
| gDNA | NOVA-5188-01-NEXTflex-Rapid-DNA-Seq-kit-2.0_v19.12_v5 | AGILENT SureSelect Clinical Research Exome v3 (CREV3) | NEXTFLEX™ Pre- & Post- Capture Combo Kit- Set A Manual (2613_0) | Illumina NovaSeq 6000 |
| cfDNA | NOVA-5150-01_NEXTFLEX_Cell_Free_DNA-Seq_Kit_19.02-002 | AGILENT SureSelect Clinical Research Exome v3 (CREV3) | NEXTFLEX™ Pre- & Post- Capture Combo Kit- Set A Manual (2613_0) | Illumina NovaSeq 6000 |
Table 2. Details of the NGS workflow for relevant sequenced products.
One batch per product was sequenced, the results for endogenous variants cannot necessarily be claimed to be validated. However, the high coverage (>500x) of the data provides confidence in our results.
What information is provided on the sequenced products?
Each product has two VCF files (aligned against GRCh37/hg19 or GRCh38/hg38) which contain the following variant information:
- Chromosome location
- Gene name
- Genomic coordinates of the variants (aligned against GRCh37/hg19 or GRCh38/hg38)
- dbSNP ID
- Nucleotide substitution
- Read depth
VCFs are large (>200MB) text-based files that can be opened using your favorite bioinformatic analysis software (e.g. bedtools, VCFtools, etc.). Alternatively, they can be unzipped using a tool like 7-Zip and then opened using a text editor (e.g. Sublime Text, Notepad++, etc.).
I have different variants detected than those found in the VCF files. What could be causing this?
We expect there to be slight batch differences or differences in the formats of the products manufactured using the same cell line blend due to biological and technical variabilities. Variant detection is also dependent on the NGS chemistry and the bioinformatics pipeline used.
Supporting data analysis
Detecting SNPs vs MNPs
A multiple nucleotide polymorphism (MNPs) is characterized by two or more SNPs occurring right next to each other, these variants are sometimes called MNPs.
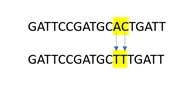
We understand researchers may call out MNP’s in their sequencing. However, the software used for data analysis calls out each variant as a SNP and we have not listed them as MNP variants. We have not verified the co-occurrence of the consecutive SNPs so it is possible that there may be MNP variants present, depending on the Allele Frequency (AF) of each SNP and their simultaneous occurrence. However, the genomic coordinate and the read depth of the individual SNP information is available in these data which can help speculate on the nature of the variant (SNP or MNP).
Detecting true positives vs false negatives
With the availability of WES data for Horizon products, confirming true positives and false negatives is facilitated. Our WES data is comprehensive compared to the targeted panel sequencing that others may use. We can only confidently call those variants that have been validated via ddPCR (available on each product page).
The presence of an endogenous variant can be confirmed with the WES data provided. A high coverage dataset gives us confidence in confirming the presence of variants. However, we cannot verify the absence of a variant that was not detected as it could be either a true negative or missed detection due to the assay design or analysis method used.
Based on the assay type, assay design and the data analysis method used by the customer, there can be differences in the variants called or the %AF. We have used Illumina NovaSeq 6000, where the kit and data analysis information will be provided on the exome sequencing page.
Can we get sequencing data for other formats?
For products where Horizon sells multiple formats, e.g Structural multiplex in gDNA (HD753), cfDNA (HD786) and FFPE (HD789), we purposely chose the gDNA format to be sequenced. This is because cfDNA and FFPE products may show artifacts due to sonication or fixation. Sequencing gDNA ensures the highest confidence variant calling.
cfDNA product selection
The two cfDNA products sequenced are from the HD780 Multiplex cfDNA set. The 5% cfDNA product (HD777) was chosen so that the claimed variants can be confidently verified. The data from the wild type (WT) product (HD776) will also verify the true negatives as well as provide information on other endogenous variants.
Written by Bernice Freeman & Jesse Stombaugh

 Jesse is a Senior Bioinformatics Scientist in the Bioinformatics team at Horizon Discovery, US. He obtained his PhD from Bowling Green State University, where he focused on RNA 3D structure prediction. At Horizon, he specializes in developing and scaling software solutions for processing and analyzing NGS data. These solutions are used for developing new products and assessing the quality of our current products.
Jesse is a Senior Bioinformatics Scientist in the Bioinformatics team at Horizon Discovery, US. He obtained his PhD from Bowling Green State University, where he focused on RNA 3D structure prediction. At Horizon, he specializes in developing and scaling software solutions for processing and analyzing NGS data. These solutions are used for developing new products and assessing the quality of our current products.
