Gene overexpression experiments using CRISPRa are a powerful means of achieving robust and specific target up-regulation of endogenous product, without permanent alterations to DNA.
CRISPRa experiments require in-cell delivery (or residence) of an engineered nuclease-deactivated Cas9 (dCas9) fused to a trio of transcriptional activators (VP64, p65 and Rta, or “VPR”) and a predesigned guide RNA that targets near the gene promotor region. Several options and combinations of installing dCas-VPR and the guide RNA into cells exist, each with implications for specific cell types, the scientific question being asked and desired effect duration. A non-exhaustive list of options for delivering CRISPRa components and considerations include:
Obtaining, or generating a cell line that stably expresses both dCas9-VPR and guide RNA
- Extended activation enables long-term overexpression phenotype
 Stable expression of components preclude ability to control activation timing
Stable expression of components preclude ability to control activation timing- Limited cell line options and difficult-to-transduce (by lentivirus) cells are excluded
Transient transfection / electroporation of stable cell line with synthetic guide RNA
- Adjusting delivery timing of guide RNA enables controlled initiation of CRISPRa effect
- Limited to cell lines that can handle transduction by lentivirusGuide RNA eventually degrades, terminating CRISPRa effect
Transient transfection of dCas9-VPR-encoding mRNA and synthetic guide RNA (see Fig 1)
- No exogenous DNA introduced and no permanent alterations to genome
- Precise control over expression of all CRISPRa components
- mRNA and guide RNA eventually degrade, ensuring a short time point assay
- Good option for difficult-to-transduce cells
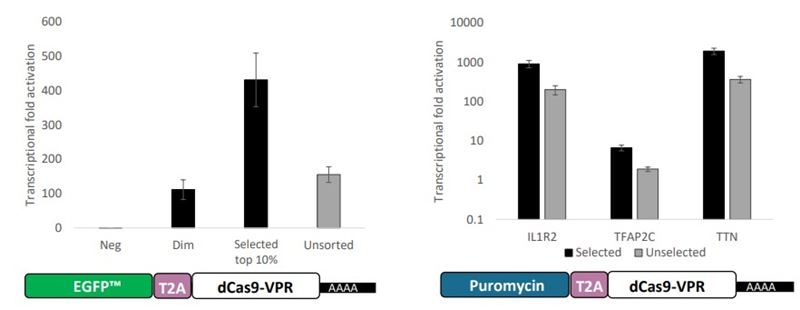
The final CRISPRa overexpression phenomenon using a combination of dCas9-VPR mRNA and synthetic guide RNA is temporally governed by a mixture of cellular kinetics of translation initiation, RNP formation and RNA decay (mRNA and guide RNA may decay at different rates). To activate a target gene, guide RNA must complex with dCas9-VPR protein in the nucleus. For this to happen, the mRNA must be translated in the cytoplasm, complex with the guide RNA, and make its way to the genomic target. Since cytoplasmic RNA will eventually decay at a rate contingent upon its makeup and chemical modifications, the mRNA and guide RNA will likely encounter different cellular lifetimes. As such, it is difficult to predict the advent and decline of gene activation in these complicated transient experiments. Further, since clonal dCas9-VPR expressing cell lines are not employed, homogeneity of effect (population penetration) is unknown (although enrichment is possible using mRNA with co-expressed fluorescent markers or antibiotic resistance [Fig 2]).
To this end, we developed resources for those interested in pursuing such gene activation experiments using mRNA and synthetic guide RNA. Details on these application notes are as follows:
Duration of CRISPRa-Guided Target Overexpression Using dCas9-VPR-EGFP mRNA. Here, we demonstrate the timeline and kinetics of dCas9-VPR translation in a model cell line, when co-transfected with dCas9-VPR-EGFP mRNA and target guide RNA. In the same cells, we also demonstrate the timeline of target gene mRNA and protein overexpression. We find that such co-transfection experiments enable the controllable, rapid, and robust overexpression of target gene products within two days of introduction. Importantly, for the targets studied, expression returned to basal levels within 4-7 days, making it possible to analyze cells before, during and after target overexpression. Read the full application note below.
Population Penetrance of CRISPRa-Guided Target Overexpression Using dCas9-VPR-EGFP mRNA. Using the same targets and cell line as above, we assess the penetrance, or degree of cells exhibiting the phenomenon being studied, in CRISPRa experiments. Notably, for the two targets under study, there was nearly complete phenotype penetrance, which was in-line for parallel experiments where dCas9-VPR was stably produced. Read the full application note here.

