CRISPRmod (CRISPRi & CRISPRa) experiments require two components be delivered and/or expressed in the cell for efficient transcriptional modulation: a targeted guide RNA and a nuclease-deactivated Cas9 (dCas9) fused to either transcriptional repressors (SALL1 and SDS3) or activators (VPR). Insufficient amounts of either component will result in inefficient knockdown/activation of the target gene, so proper controls should be used to ensure success.
Controls should be used for:
- Optimizing synthetic guide RNA transfection conditions
- Determining the optimal promoter for driving lentiviral dCas9 expression
- Identifying optimal co-transfection conditions with dCas9 mRNA
- Ensuring experimental consistency and controlling for any possible background effects
CRISPRi controls for loss-of-function experiments
To establish optimal experimental conditions and ensure ongoing successful gene repression, it is recommended to use a specific CRISPRi positive control for a characterized gene target. The CRISPRi positive controls target SEL1L or PPIB genes.
Non-targeting (negative) controls are guide RNAs with no complementary target in the annotated human genome, so the baseline expression of any given gene should not be altered. mRNA and protein levels may change over time in CRISPRmod experiments; this should be taken into consideration when detecting expression levels. Use qPCR and Western blotting to determine changes in mRNA and protein levels compared to untreated and non-targeting controls to observe gene modulation effects. Read more about CRISPRi applications.
CRISPRa controls
To establish optimal experimental conditions and ensure ongoing successful activation, it is recommended to use a species-specific CRISPRa positive control for a characterized gene target. The CRISPRa positive controls target human or mouse Titin (TTN) or POU class 5 homeobox 1 (POU5F1) genes. Read more about CRISPRa applications.
Robust gene knockdown with CRISPRi synthetic sgRNA positive controls

K562 and Jurkat cells were nucleofected with dCas9-KRAB or dCas9-SALL1-SDS3 mRNA (2 µg), and pooled CRISPRi synthetic sgRNAs (5 µM) targeting PPIB and SEL1L via a Lonza 96-well Shuttle system. WTC-11 hiPS cells were nucleofected with dCas9-KRAB or dCas9-SALL1-SDS3 mRNA (1 µg) and pooled CRISPRi synthetic sgRNA (3 µM) targeting PPPIB or SEL1L via a Lonza 96-well Shuttle system. Cells were harvested 72 hours post-nucleofection. Total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative gene expression for each target gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).
Efficient transcriptional gene activation with CRISPRa synthetic crRNA:tracrRNA in dCas9-VPR stable cells
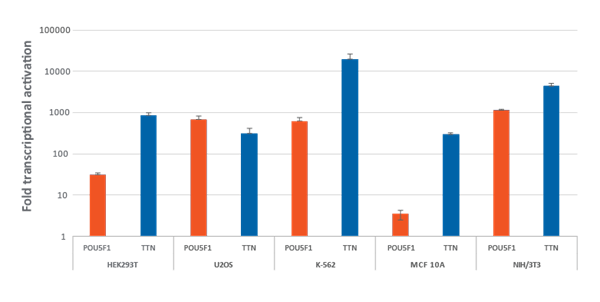
Efficient transcriptional gene activation with CRISPRa synthetic crRNA:tracrRNA in dCas9-VPR stable cells HEK293T, U2OS, MCF 10A, NIH/3T3 stably expressing integrated dCas9-VPR were plated at 10,000 cells/well and transfected using DharmaFECT Transfection Reagents with CRISPRa synthetic crRNA:tracrRNA (25 nM) targeting POU5F1 and TTN. K562 cells were electroporated with CRISPRa synthetic crRNA:tracrRNA (400 nM) targeting POU5F1 and TTN. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ΔΔCq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
Efficient transcriptional gene activation with lentiviral CRISPRa sgRNA in dCas9-VPR stable cells
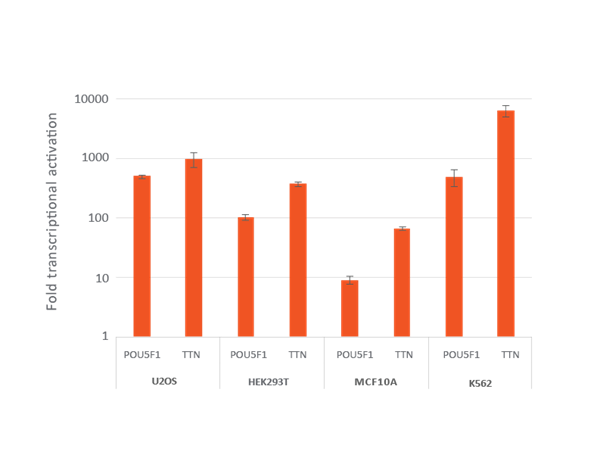
U2OS, HEK293T, MCF 10A and K562 stably expressing integrated dCas9-VPR were plated at 10,000 cells/well and transduced with CRISPRa sgRNA lentiviral particles targeting POU5F1 or TTN at a MOI of 0.3 to obtain cells with a single integrant. Cells were selected with 2 µg/mL puromycin for 4 days prior to analysis with RT-qPCR. The relative expression of each gene was calculated with the ΔΔCq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
Order CRISPRi Controls
CRISPRi synthetic sgRNA positive controls
Pooled or individual sgRNA controls for assessment of optimal experimental conditions for gene repression
CRISPRi synthetic sgRNA non-targeting controls
Non-targeting controls to evaluate baseline cellular responses to CRISPRi components in the absence of gene target-specific sgRNA
CRISPRi lentiviral sgRNA positive controls
Control sgRNAs targeting well-characterized genes to determine the effectiveness of your experimental conditions for maximum repression
CRISPRi lentiviral sgRNA non-targeting controls
Non-targeting controls to evaluate baseline cellular responses to CRISPRi components in the absence of target-specific sgRNA
Order CRISPRa Controls
CRISPRa synthetic crRNA positive controls
Pooled or individual crRNA controls for assessment of optimal experimental conditions for gene activation
CRISPRa synthetic crRNA non-targeting controls
Non-targeting controls to evaluate baseline cellular responses to CRISPRa components in the absence of gene target-specific crRNA
CRISPRa lentiviral sgRNA positive controls
Control sgRNAs targeting well-characterized genes to determine the effectiveness of your experimental conditions for maximum activation
CRISPRa lentiviral sgRNA non-targeting controls
Non-targeting controls to evaluate baseline cellular responses to CRISPRa components in the absence of target-specific sgRNA
Order All-in-one Controls
CRISPRi All-in-one Lentiviral sgRNA Positive Controls
All-in-one lentiviral sgRNAs targeting well-characterized genes to determine the effectiveness of your experimental conditions for maximum transcriptional repression.
CRISPRi All-in-one Lentiviral sgRNA Non-targeting Controls
Bioinformatically designed All-in-one Non-targeting sgRNA controls to evaluate baseline cellular responses to CRISPRi components in the absence of target-specific sgRNA.
CRISPRa All-in-one Lentiviral sgRNA Positive Controls
All-in-one lentiviral sgRNAs targeting well-characterized genes to determine the effectiveness of your experimental conditions for maximum activation.
CRISPRa All-in-one Lentiviral sgRNA Non-targeting Controls
Bioinformatically designed All-in-one Non-targeting sgRNA controls to evaluate baseline cellular responses to CRISPRa components in the absence of target-specific sgRNA.
