Reference standards can serve as a control for a diagnostic assay, the whole workflow, and during assay development to understand the sensitivity of an assay. This article briefly describes how our reference standards are produced and the quality control measures to ensure the standards are reliable from batch to batch. Understanding these technical points provides an insight into how a reference standard from Horizon will mimic patient samples and how it can be used as a control (Figure 1).

How standards mimic patient sample workflow

Standards for different parts of diagnostic workflow
| Reference standard formats | |
|---|---|
| gDNA | Genomic DNA from cell lysates |
| cfDNA | DNA in Buffer or synthetic plasma |
| fcDNA | Formalin Compromised DNA |
| FFPE DNA | Formalin-Fixed and Paraffin-Embedded DNA |
| FFPE RNA | Formalin-Fixed and Paraffin-Embedded RNA |
Regardless of format, Horizon’s reference standards are produced as cell-line derived reference standards to resemble the genomic complexity of a patient’s sample. Cells with genetic variations such as insertions and deletions (INDELs), fusions, single nucleotide variants (SNVs) and copy number variants (CNVs) are blended to produce our reference standards. These variants have been constructed from both existing cell lines harboring the variant of interest and by engineering mutations into cell lines using genetic engineering techniques such as rAAV and CRISPR. Cell lines are blended to create variants at specified frequencies and then offered in various formats (see Table 1) to mimic diagnostic samples and for use in a variety of assays. In contrast, competitor’s reference standards are often produced by using a characterized whole-genome extract (such as NIST’s GIAB cell lines) with synthetic DNA variants spiked into a pool. Horizon’s reference standards use mutations encoded within the genomic DNA of the cell line to better mimic patient samples. This unique strategy allows the mutation to be present in the context of the whole genome at the correct genetic loci and surrounding genetic environment. Customers have identified this as an important factor when performing subsequent bioinformatic analysis and NGS workflow optimization when using Horizon standards compared to synthetic standards.
We perform quality control analysis for the variants specified for each reference standard batch to ensure consistency, but more variants may exist in a standard than those guaranteed due to the complexity of a whole-genome product. A multiplex reference standard can act as multiple controls in one sample freeing up more room for patient samples in an assay run. Our family of Oncospan reference standards comes with batch-specific bioinformatic data available on purchase, allowing researchers to know other variants and backgrounds that are present in the standard.
Horizon’s reference standards have variants at varying allelic frequencies that mimic the complexity of biological samples seen in-clinic. Typically, an allele can be found at 50% or 100% allelic frequency depending if the allele is heterozygous or homozygous in a cell line. Blending a variant cell line with additional cell lines that harbor different genetically engineered or endogenous mutations generates reference standards with different allelic frequencies. This cell line blend also allows Horizon’s reference standards to mimic the heterogeneity in biological diagnostic samples (Figure 3).
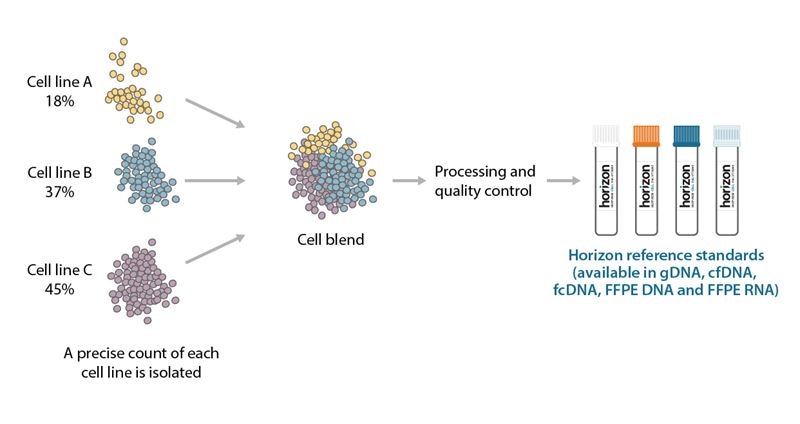
Cell blending strategy
Horizon offers both singleplex reference standards, that allow control for a single allele/variant of interest, and multiplex reference standards that can act as a control for multiple alleles/variants at differing allelic frequencies. Complexed, characterized reference standards at varying allelic frequencies allow researchers to know the limits of detection of their assay, have confidence in their results, and since multiple controls can be combined using our multiplex standards, they allow labs to run more patient samples.
Horizon performs quality control on each batch of reference standards to ensure variants are found at precise allelic frequencies by digital droplet PCR (ddPCR). The sensitive ddPCR analysis provides a consistent benchmark for your experiments. Detection of variants using different assays, such as NGS may vary based on many factors, some of which are discussed here. Results from our analysis are provided online in the batch-specific certificate of analysis (CoA) found in the ‘Resources’ tab on each product page. The quantity and integrity of each reference standard is checked by quality control measures to ensure a high-quality genetic material (Table 2).
|
Reference standard quality control |
||
|---|---|---|
| Reference standard(s) | Quantification Method | Assay for integrity |
| gDNA | Spectrophotometry at A260 - Nanodrop | Agarose Gel Electrophoresis |
| cfDNA, fcDNA | Fluorescent DNA detection – Thermo Qubit® dsDNA BR (broad range) assay | Tapestation |
| FFPE DNA | Fluorescent DNA detection - Promega QuantiFluor® dsDNA assay | Agarose gel electrophoresis |
| FFPE RNA | Fluorescent RNA detection – Thermo Qubit® RNA HS assay | FFPE RNA tested for amplifiability by end-point RT-PCR |
Reference standards can allow researchers to have confidence in their assay and act as a process control to understand problems in a diagnostic workflow. Horizon’s offering of reference standards is curated to mimic the complex nature of biological patient samples and provide reliable, reproducible, and affordable options. Should you have any questions about our reference standards or need help finding the standard for your diagnostic assay please reach out to us our Scientific Support team.
Author: Christopher Wrobel Ph.D. | Scientific Support Specialist
Order Horizon reference standards
Browse offerings
Learn more
- The five quality control metrics every NGS user should know Blog article
- Using reference standards in the development of new liquid biopsy platforms – Blog article
- Reference standard formats commutable with tumor biopsy and liquid biopsy samples for cancer genomic profiling – Poster
