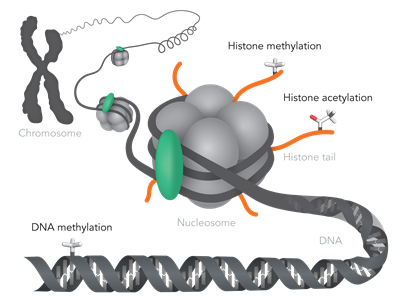
Epigenetic Marks (Markers)
The most important epigenetic marks include DNA methylation, histone methylation and histone acetylation.
All of these are reversible, i.e. the cell harbors a repertoire of enzymes that add or remove such marks to a specific genomic locus.
Enzymes that add marks are referred to as epigenetic writers (e.g. DNA methyl transferases or histone acetyl transferases) and those that remove marks are known as epigenetic erasers (e.g. histone demethylases or histone deacetylases).
Epigenetic Modification
Epigenetic marks are read-out by a specific subset of proteins referred to epigenetic readers (e.g. bromodomain-containing genes). Some examples of epigenetic mechanisms include; maternal effects, X-chromosome inactivation and imprinting amongst many others.
Explore our popular knockout cell lines for Epigenetics
Order products
Human knockout HAP-1 cells
The single largest bank of isogenic cell lines with over 7,500 cell lines to choose from and trusted by academia, biotech, and pharma research labs.
Cancer-related cell lines
Choose from over 300 knock-in and knockout cell line models in many standard cancer cell lines such as DLD1, MCF10A, and HCT116.
Cas9 Stable Cell Lines
Simplify gene editing experiments with stably expressing Cas9 cell lines
CRISPRmod CRISPRa dCas9-VPR Stable Cell Lines
Streamline CRISPR activation experiments with stably expressing dCas9-VPR cell lines
