Add any of our CRISPR portfolio reagents (CRISPRko, CRISPRa, CRISPRi) to your cart to see new pricing (up to 56% off) now including All-in-one lentiviral sgRNA formats in 2026
Cas9 nuclease solutions for every CRISPR gene editing workflow
Horizon offers a variety of Cas9 nuclease solutions for CRISPR-based gene editing experiments.
Learn more about the Edit-R™ CRISPR-Cas9 system and gene editing applications.
Cas9-expressing stable cell lines for streamlined gene editing
Shorten your next gene editing workflow with "CRISPR-ready" Cas9-expressing stable cells in a variety of popular cell types.
Fluorescent Cas9 reagents for optimization and enrichment
Simplify enrichment of Cas9 expressing cells with lentiviral or mRNA reagents that co-express either mKate2 or TurboGFP™ with Cas9.
Transient Cas9 products for DNA-free gene knockout
Reduce off-targets and protect cells from potential unwanted cellular responses with mRNA or protein reagents that permit transient Cas9 expression.
Lentiviral Cas9 for easy delivery into any cell type
Eliminate the need for cytotoxic transfection or electroporation steps with constitutive or Strict-R inducible lentiviral Cas9 reagents.
Cas9 nuclease selection guide
Determining the most appropriate Cas9 nuclease reagent for your experiment is dependent on the particular application or cell type.
See the table below to determine the best format for your experiment.
| Cas9 protein | Cas9 mRNA | Lentiviral Cas9 particles | |
|---|---|---|---|
| DNA-free, transient expression | ✔ | ✔ | |
| Co-electroporate with synthetic guide RNA | ✔ | ✔ | |
| Co-transfect with synthetic guide RNA | ✔ | ✔ | |
| Enrich population with FACS | ✔ | ✔ | |
| Enrich population with resistance marker | ✔ | ||
| Inducible expression | ✔ | ||
| *NEW* Strict-R inducible expression | ✔ | ||
| Create stable cell lines | ✔ | ||
| Lentiviral transduction for cells that are difficult to transfect | ✔ |
Order Cas9 stable cells
Cas9-expressing cells
Cas9-expressing stable cells in a variety of popular cell types - simply introduce Edit-R CRISPR guide RNA for targeted gene knockout.
Order transient Cas9 nuclease
Cas9 mRNA
Purified Cas9 mRNA for transient Cas9 nuclease expression with fluorescent options for sorting, enrichment & visualization.
New! Edit-R Cas9 Protein Hybrid NLS
Purified ready-to-use Cas9 protein with a new and improved NLS composition enabling more efficient nuclear delivery for rapid RNP workflows.
Cas9 protein NLS
Purified ready-to-use Cas9 protein for rapid RNP workflows.
Order vector-based Cas9
Lentiviral Cas9
Purified lentiviral particles or plasmid DNA for generating stable Cas9-expressing cells. Inducible promoter options available.
Strict-R inducible Cas9 lentiviral system
Lentiviral Cas9 system for dual controlled gene knockout in diverse cell types.
Cas9 mRNA data
Efficient indel formation using Edit-R synthetic sgRNA in Cas9-expressing stable cell lines
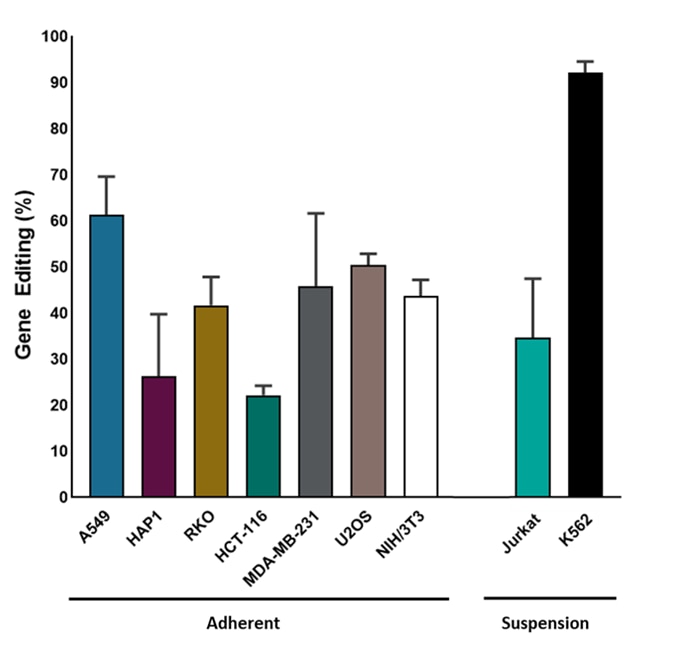
Adherent Cas9 Stable Cell Lines were plated at 10,000 cells/well and reverse-transfected using DharmaFECT1 or DharmaFECT4 Transfection Reagent with synthetic sgRNA (25 nM) targeting PPIB (Cat # U-009000-01-XX). Cells were harvested 72 hours post-transfection, and the relative efficiency of indel formation was assessed using trace decomposition (TIDE) compared to a non-targeting control (NTC; Cat # U-009501-01-XX). Suspension Cas9 Stable Cell Lines were electroporated using 100,000 cells/well, and Lonza buffer SE or SF, respectively, with synthetic sgRNA (5 µM) targeting PPIB (Cat # U-009000-01-XX). Cells were harvested 72 hours post-transfection and the relative efficiency of indel formation was assessed using trace decomposition (TIDE) compared ion to a non-targeting control (NTC; Cat # U-009501-01-XX).
Fluorescent Cas9 nuclease mRNA maintains editing efficiency
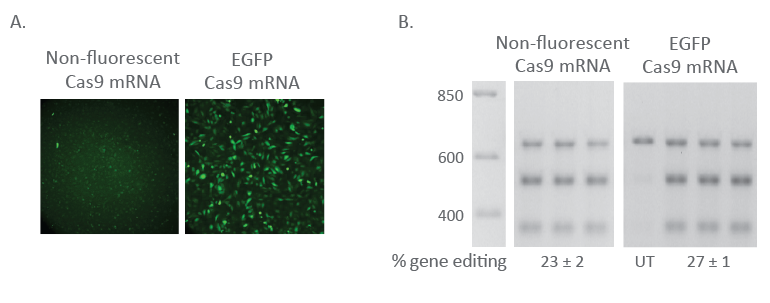
Fluorescent Cas9 mRNA U2OS cells were plated at 10,000 cells/well in a black 96-well plate 24 hours before transfection. At the time of transfection, EGFP Cas9 nuclease mRNA (200 ng, Cat #CAS11860) or non-fluorescent Cas9 Nuclease mRNA (200 ng, Cat #CAS11195) was co-transfected with Edit-R PPIB Synthetic crRNA Control (25 nM, Cat #U-007000-01-05) and tracrRNA (25 nM, Cat# U-002005-05) using DharmaFECT™ Duo transfection reagent (0.3 µL/well). After 24 hours, cells were imaged for EGFP fluorescence using the Nikon Eclipse Ti-S/L100 inverted microscope (A). At 72 hours after transfection, cells were harvested and gene editing was measured through a DNA mismatch detection assay (B).
Fluorescent Cas9 nuclease mRNA indicates optimal transfection conditions
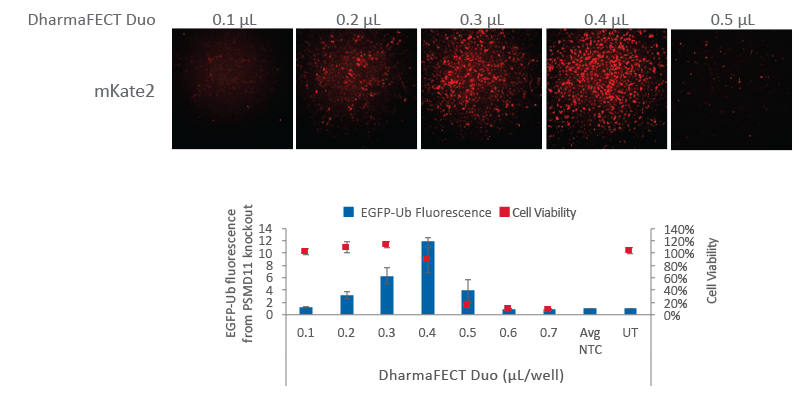
U2OS cells expressing a Ubiquitin-tagged EGFP were plated at 10,000 cells/well in a black 96-well plate and co-transfected with Cas9 mRNA (200 ng, Cat #CAS11859) and synthetic tracrRNA (25 nM, Cat# U-002005-05) targeting PSMD11 (25 nM, Cat #CM-085161-03), a known proteasome component, or a Non-targeting control (NTC, Cat #U-007501-xx). Increasing amounts of DharmaFECT Duo Transfection Reagent were tested per well (0.1 µL to 0.7 µL). After 24 hours, cells were imaged for mKate2 fluorescence using the In Cell Analyzer 2200. At 72 hours after transfection, cells were imaged for EGFP fluorescence resultant from proteasome knockout. Based on mKate2 fluorescence, optimal transfection conditions occur at 0.3-0.4 µL of DharmaFECT Duo per well, which corresponds to an increased phenotypic effect. Above 0.4 µL of DharmaFECT Duo per well, significant cell death is observed (graph, red boxes). UT = untransfected sample.
Cas9 protein data
Nucleofection of CD4+ primary human T-cells with Edit-R Cas9 Protein Hybrid NLS RNPs
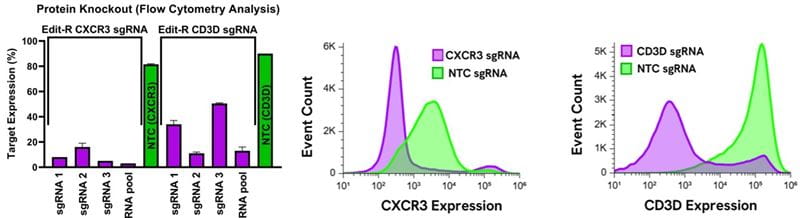
CD4+ cells were stimulated with CD3/CD28 T-cell activation beads (BioLegend) at a 1:1 bead:T-cell ratio and grown for 8 days with 30 U/mL of IL-2. The Lonza 4D shuttle system was used to nucleofect RNPs composed of Edit-R Cas9 Protein Hybrid NLS and Edit-R synthetic guide RNA individual and target pools for CXCR3 (Gene ID: 2833) and CD3D (Gene ID: 915). Functional knockout was measured by flow cytometry analysis of target prevalence for CXCR3 (Clone ID: G025H7) and CD3D (Clone ID: OKT3) (BioLegend). Representative flow cytometry analysis plots of CD4+ T-cells shown after target knockdown by RNP nucleofection.
Lentiviral Cas9 data
Fluorescent lentiviral Cas9 nuclease enables enrichment for high expression and improved CRISPR-Cas9 gene editing efficiency
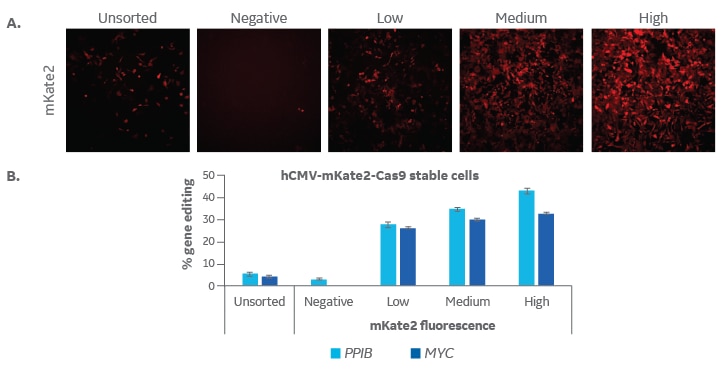
U2OS cells were transduced at low multiplicity of infection (MOI 0.3) with Edit-R Lentiviral hCMV mKate2-Cas9 Nuclease particles (Cat #VCAS11869) so that transduced cells would have only one integration of Cas9. Cells were expanded for fluorescence activated cell sorting (FACS) where populations were sorted into negative, low, medium and high mKate2 fluorescence. These subpopulations were expanded and then plated at 10,000 cells/well in a 96-well plate. One day later, cells were transfected with tracrRNA (25 nM, Cat #U-002005-xx) and Edit-R PPIB Synthetic crRNA Control (Cat #U-007501-xx) or Edit-R MYC Predesign crRNA (Cat #CM-003282-01) using DharmaFECT 1 transfection reagent (0.3 µL/well, Cat #T-2001-01). After 72 hours, cells were imaged for mKate2 fluorescence using the In Cell Analyzer 2200 (GE Healthcare; A) and then harvested for DNA mismatch detection assay to estimate gene editing (B). High mKate2 expression can be associated with the highest levels of gene editing for both PPIB- and MYC-targeting crRNAs.
Nuclease sources
"CRISPR-ready" premade Cas9-expressing cell lines
-
Cas9 expressing stable cell lines
Choose from a variety of popular cell backgrounds, then simply deliver a Edit-R CRISPR guide RNA targeted to any gene for simple loss-of-function studies.
Transient Cas9 nuclease for DNA-free workflows
-
Cas9 nuclease mRNA
Purified Cas9 mRNA for transient Cas9 nuclease expression. Fluorescent options available for sorting, enrichment and visualization of delivery. -
Edit-R Cas9 Protein Hybrid NLS
New! Purified ready-to-use Cas9 protein with a new and improved NLS composition enabling more efficient nuclear delivery for rapid RNP workflows -
Cas9 nuclease protein NLS
Purified Cas9 protein ready-to-use for DNA-free workflows.
Vector-based solutions for generating Cas9-expressing cell lines
-
Lentiviral Cas9 nuclease reagents
Purified lentiviral particles or plasmid DNA for generation of stable Cas9 nuclease-expressing cell populations. Constitutive or inducible promoter options are available.
Solutions for generating inducible Cas9 expressing cell-lines
-
Strict-R inducible Cas9 lentiviral system
Lentiviral Cas9 system for dual controlled gene knockout in diverse cell types.
CRISPR guide RNA
Edit-R synthetic guide RNA & controls
-
Synthetic sgRNA
Genome wide synthetic sgRNA designs guaranteed to edit your gene of interest. The design algorithm maximizes the potential for functional protein knockout while minimizing off-target editing through stringent specificity checks.
-
Synthetic sgRNA positive controls and detection primers
Species-specific synthetic sgRNAs targeting well-characterized genes, as well as mismatch detection assay primers, to determine the effectiveness of your gene editing conditions for maximal efficiency.
-
Synthetic sgRNA non-targeting controls
Non-targeting controls to evaluate cellular responses to CRISPR-Cas9 components in the absence of gene target-specific sgRNA.
-
-
Synthetic crRNA
Guaranteed to edit your target! Algorithm-optimized crRNA for genome-wide coverage of human and mouse genes. Modifications for nuclease resistance improve DNA-free editing.
-
Synthetic tracrRNA
Trans-activating CRISPR RNA (tracrRNA) is required for use with Edit-R synthetic crRNA to form the complex that programs Cas9 nuclease.
Edit-R lentiviral sgRNA & controls
-
All-in-one lentiviral sgRNA + Cas9
Combined Cas9 and sgRNA expression for efficient gene knockout & unparalleled specificity; available as glycerol stocks and high-titer purified particles.
-
All-in-one lentiviral sgRNA positive controls and kits
All-in-one lentiviral sgRNA controls to verify DNA double-strand breaks and gene editing efficiencies.
-
All-in-one lentiviral sgRNA non-targeting controls
All-in-one lentiviral sgRNA constructs bioinformatically designed to not target any gene in human or mouse genomes.
-
Lentiviral sgRNA
Guaranteed to edit your target, algorithm-optimized sgRNA for genome-wide coverage of human or mouse genes. Provided as high-titer lentiviral particles or glycerol stocks.
-
Lentiviral sgRNA positive controls
Species-specific sgRNAs targeting well-characterized genes to determine the effectiveness of your gene editing conditions for maximum efficiency.
-
Lentiviral sgRNA non-targeting controls
Non-targeting controls to evaluate cellular responses to CRISPR-Cas9 components in the absence of gene-specific sgRNA.
Custom guide RNA design
-
CRISPR Design Tool
Place a custom guide RNA order, or design and order your own synthetic sgRNA, crRNA, or lentiviral sgRNA with our easy-to-use interface.
Edit-R HDR donor template design, ordering tools & kits
-
HDR Donor Designer - oligo
Design and order a single-strand DNA donor (≤ 150 nt) for insertion, deletion, or other alteration.
-
HDR Donor Designer - plasmid
Design and order a plasmid DNA donor kit for insertion of a mKate2 or EGFP fluorescent marker, or other a custom insert.
-
HDR plasmid donor kits
Rapidly and easily assemble a plasmid donor for HDR
-
HDR plasmid donor components
Quickly and efficiently build a HDR donor plasmid
-
HDR plasmid donor primers
PCR components for Edit-R Plasmid Donor Kits
The Edit-R guarantee
We guarantee that EVERY predesigned guide RNA will provide successful editing at the target site when delivered as described in the Edit-R technical manuals.
The Edit-R guide RNA guarantee is valid when used with any wild type S. pyogenes Cas9 nuclease, including mRNA, expression plasmid, protein, or stable Cas9 expression, and Edit-R crRNAs must be used with Edit-R tracrRNA for the guarantee to apply.
Analysis of editing of the treated cell population must be shown using a T7EI or Surveyor mismatch detection assay. If successful editing is not observed for a predesigned Edit-R guide RNA while an appropriate side-by-side Edit-R positive control is successful, a one-time replacement of a different predesigned Edit-R guide RNA of the same format and quantity will be provided at no cost.
A replacement will only be approved upon discussion with our Scientific Support team.
Successful editing at the DNA level does not always lead to functional gene knockout; it is recommended to test multiple guide RNAs to determine the most effective guide RNA for knockout of your target gene.
This guarantee does not extend to any accompanying experimental costs, does not apply to guide RNAs ordered via the CRISPR Design Tool, and will not be extended to the replacement guide RNA.
