
Presenting a CRISPR interference screening service (CRISPRi) that provides new opportunities for your research through loss-of-function gene modulation
This novel all-in-one human whole-genome CRISPRi platform is suited to study druggability and can effectively identify which genes are resistant or sensitive to your potential therapeutic. Utilize loss-of-function screening with the accuracy of the CRISPR guide system that’s been optimized and run by CRISPR screening experts
CRISPRi screening service
CRISPR Interference (CRISPRi) screening enables the study of reduced gene expression, rather than completely eliminating the target gene. By utilizing dead Cas9 (dCas9), which retains the ability to bind to DNA without being catalytically active, the modulation of gene expression can be achieved to provide a highly precise and sensitive loss-of-function screening solutions.
CRISPRi applications
Enabling you to:
- Model the effect of druggability more closely
- Study the effect of repressing essential genes and genes in amplified loci
- Simulate hypomorphic mutations and partial loss-of-function
- Target non-protein coding regions (e.g. lncRNA)
- Validate hits from a KO or RNAi screen with an orthologous tool
- Combine with KO or CRISPRa screening for high sensitivity screening
Horizon’s CRISPRi platform
Our CRISPR interference screening platforms are based on our industry leading highly sophisticated CRISPR KO screening platform. This CRISPRi screening platform has been optimized for more than 10 years with internal leading experts to provide a robust screening solution that offers the flexibility to fit your exact needs. The service offers:
- Both pooled lentiviral and arrayed synthetic reagent options
- Cell line(s) from a pre-optimized cell line list or cell line evaluation prior to the screen
- Human whole-genome CRISPRi library or custom designed sub-library
- Customizable screen design and readout options
- Next-generation sequencing and bioinformatics analysis with hit nominations
- Project timeline of 12-24 weeks
CRISPRi platform deliverables:
- A list of genes of which inhibition/activation alters response to the compound of interest
- A final report containing experimental design, all raw & analyzed data and conclusions of the study
CRISPRi case study data
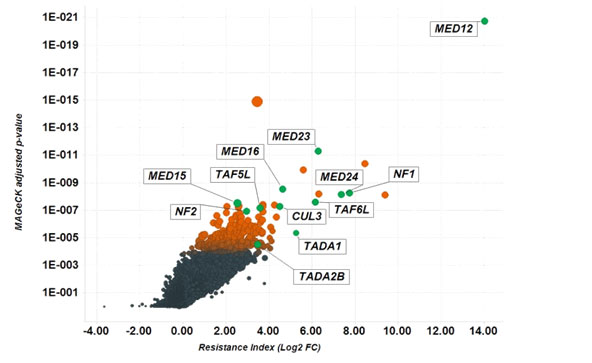
Full screen data showing log2 fold enrichment of each gene and associated p-values. Highlighted hits have been previously identified and validated by CRISPR knock-out screening.
