In early-stage pharmaceutical research, prompt identification of genes crucial to T cell function is essential for effectively choosing targets for immune-related disorders. Obtain precise insights and evaluate the functional significance of gene targets within a biologically meaningful context with a pooled T cell screen.
Pooled screening, a cost-effective and high-throughput strategy, delivers significant potential in swiftly identifying prospective therapeutic targets. Our pooled T cell CRISPR KO screens alleviates challenges in cost, scope, speed, reproducibility, and precision in gene editing. Propel your comprehension of T cell biology and discover potential therapeutic targets.
What is a pooled CRISPR KO screening in T cells?
It is a powerful method for systematically investigating gene function within a pool of T cells. The process involves designing sgRNA libraries that target a broad collection of genes and transducing primary T cells with a lentiviral vector containing these sgRNAs. The Cas9 enzyme, guided by the sgRNAs, induces double-strand breaks in the target genes, leading to their knockout during cell repair. After expanding the edited T cell population, functional assays are conducted to assess the impact of gene knockouts on T cell behavior with further multiplexed readouts, including proliferation (CTG), surface and intracellular markers (FACS), protein release quantification (HTRF®), and next-generation sequencing.
CRISPR KO applications in T lymphocytes
- Identify therapeutic targets by a systematic gene exploration.
- Understand specific gene roles in a CRISPR-based functional screening immune setting.
- Assess compound mode of action in a CRISPR-modulated immune in-vitro model.
- Uncover gene-phenotype/cell interactions and study their effects on T cell behavior.
Our pooled CRISPR KO T cell screening features
The pooled approach allows simultaneous assessment of numerous genes, accelerating the discovery of novel therapeutic targets with functional relevance in immune-related disorders. This screen enhances reproducibility and reliability by leveraging CRISPR technology, enabling precise gene editing in primary T cells.
- Advanced miniaturized pipeline enabling the assessment of drug-phenotype interactions.
- Designed for the screening of bi-specifics, small molecules, or monoclonal antibodies.
- Expandable to downstream assays such as co-culture and functional read-outs, including cytokine release through HTRF® technology, NGS analytics, and bioinformatics.
CRISPR T cell pooled screening workflow
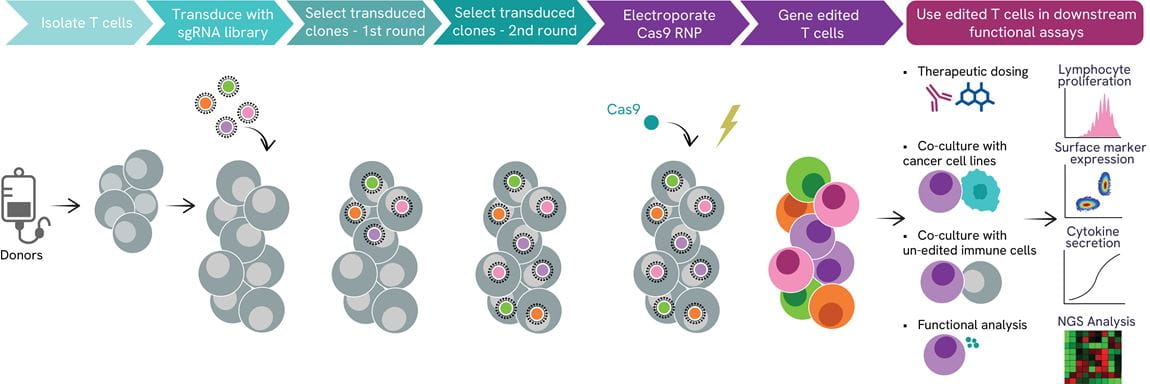
The identification of novel gene targets involved in T lymphocyte immune response by a pooled CRISPR KO screen can be achieved by our screening platform which uses isolated primary T cells, which are first transduced with a lentiviral sgRNA library ahead of the introduction of Cas9 mRNA. After editing, T cells can be co-cultured with unedited immune cells and cancer cell lines and/or challenged with a treatment of a therapeutic candidate. Proliferation, surface expression of markers of interest, cytokine release quantification, and NGS with subsequential bioinformatic analytics are integrated readouts of our pipeline.
Functional analysis by T cell activation assay of edited cells
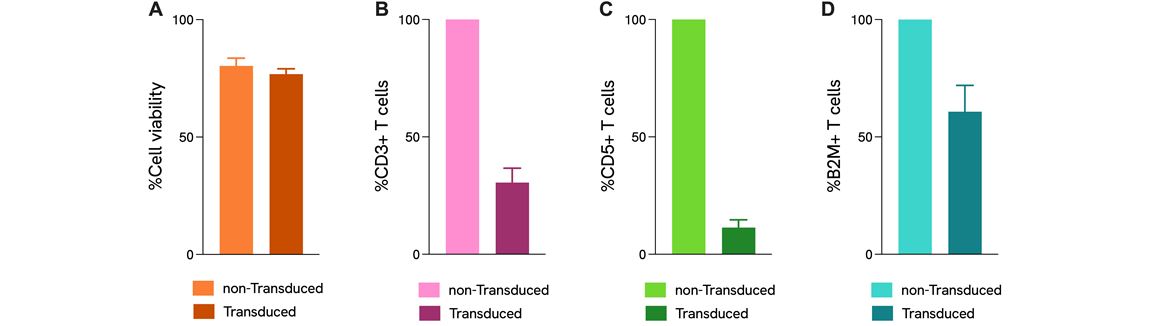
Pooled CRISPR KO in T cells. Human CD3 T cells were transduced with 9 CRISPR RNA guides targeting three genes by lentiviral delivery and electroporated with Cas9 RNP in a pooled screening format. A) Viability of edited T cells in comparison to non-transduced control. B-D) Flow cytometry measured the knock-out efficiency of CD3, CD5, and B2M, respectively, as the surface expression percentage of total live lymphocytes. Data were normalized to non-transduced control. LentiBoost-P is used as a transduction enhancer. Data is depicted from three donors.

